Figure 6.
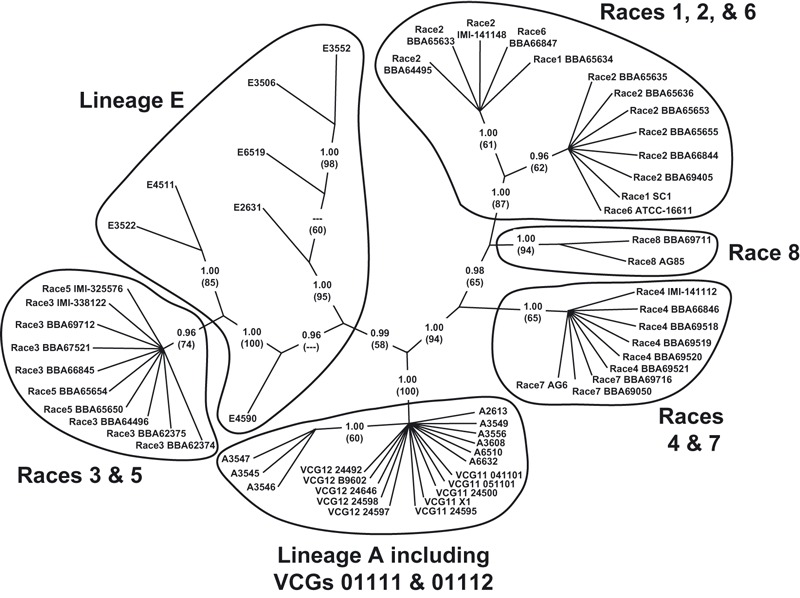
Consensus parsimony optimized phylogenetic tree based on concatenated alignment (2399 bp + four indels encoded as binary characters) of the translation elongation factor-1α (653 bp + two indels), mitochondrial small subunit rDNA (677 bp + one indel), nitrate reductase (483 bp + one indel), and phosphate permease (586 bp) gene sequences. This unrooted topology is the consensus of five equally parsimonious trees (consistency index = 0.9286; retention index = 0.9915). Posterior probabilities, estimated in a separate Bayesian analysis, are indicated for each node; bootstrap values (1000 replicates) are indicated parenthetically beneath each posterior probability.
