Figure 1.
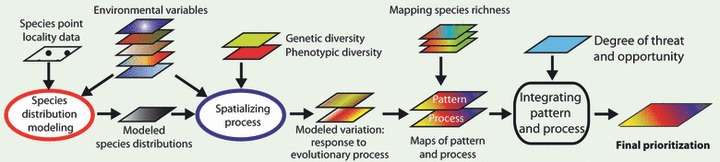
Schematic of our workflow for this paper. (i) Species distribution models for the seven target taxa were created using Maxent (Phillips et al. 2006), species-presence point localities, and a set of environmental variables. The modeled species distributions served to delimit our study area for modeling intraspecific variation. (ii) We predicted the patterns of genetic and morphological diversity using in situ data and the environmental variables in a generalized dissimilarity modeling framework (Ferrier et al. 2007). For each genetic or morphological trait, these analyses resulted in a GIS layer with values representing 50 similarity classes. These classes were separated as individual layers in ArcGIS (version 9.3) and used in (iii) subsequent ResNet reserve selection software (Sarkar et al. 2009) to prioritize areas for conservation based on genetic and morphological variation. (iv) We also used species distributions of birds, mammals, and amphibians, available from public databases, to prioritize areas for conservation based on species richness and complementarity. Finally, we combined the results from steps (iii) and (iv) and compared all results with each other and with currently protected areas.
