Abstract
Five adenovirus (Ad) gene products are required for efficient replication of co-infecting adeno-associated virus (AAV); however, the combined net enhancement by these factors is composed of both positive and negative effects. Similar to previous results with AAV Rep52, AAV2 large Rep was targeted for ubiquitination and degradation by the Ad E4orf6/E1b 55 kDa, cullin 5-containing, E3-ubiquitin ligase. Additionally, large Rep was targeted for ubiquitination via extension of ubiquitin lysine K48 and K63 both in the presence and absence of E4orf6.
Adeno-associated viruses (AAVs) are members of the genus Dependovirus within the family Parvoviridae (Berns & Giraud, 1996; Bowles et al., 2006). AAV can only achieve efficient productive replication in the presence of a larger helper virus (Bowles et al., 2006).
We have previously shown that the adenovirus (Ad)-helper functions required for AAV replication provide a surprising combination of positively and negatively acting effects, and the Ad E4orf6/E1b 55 kDa E3-ubiquitin ligase (Ad E3 Ub-ligase) can target both AAV Rep52/40 and AAV capsid proteins for ubiquitination and subsequent degradation (Nayak et al., 2008; Nayak & Pintel, 2007). The role during infection for this activity is not yet fully understood. Steady-state levels of AAV protein are not seen to be reduced during co-infection with Ad, or when supported by the full panoply of Ad-helper functions, because enhancement of AAV mRNA translation by Ad virus-associated (VA) RNA restores AAV protein to levels required for efficient infection (Nayak & Pintel, 2007). Because the AAV large Rep protein is the major viral protein required for genome replication, and because it shares approximately 2/3 of its amino acid sequence with the targeted smaller Rep proteins, we sought to determine if the large Rep protein was also a target of the Ad E4orf6/E1b 55 kDa E3 Ub-ligase. It has previously been shown that when expressed alone in cells, large Rep is a stable protein exhibiting a long half-life (Weger et al., 2004), similar to AAV/Ad co-infection. Therefore, as seen previously during our analysis of Ad effects on AAV Rep 52/40 and Cap (Nayak et al., 2008), we expected that Ad E3 Ub-ligase-targeting of Rep for degradation would be only clearly apparent in the absence of VA RNA.
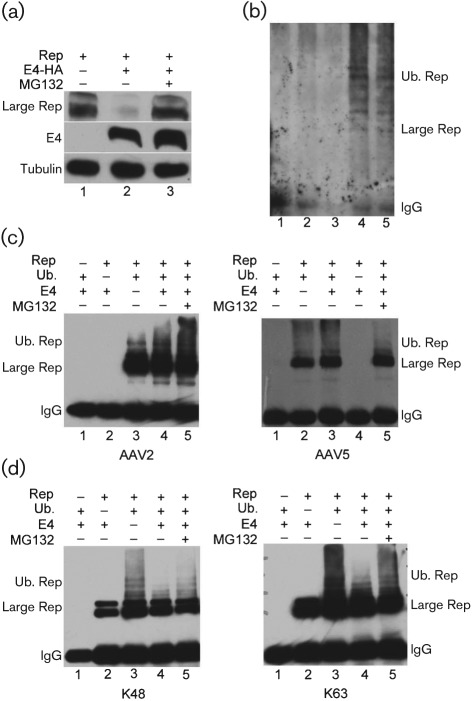
As can be seen in Fig. 1(a), accumulated levels of the large Rep protein expressed in Ad E1a- and E1b-expressing 293 cells are significantly reduced in the presence of Ad E4orf6 (compare lanes 1 and 2). Additionally, large Rep levels can be recovered in the presence of the proteasome inhibitor MG132 (Fig. 1a, lane 3), suggesting that the E4orf6-dependent degradation of large Rep is proteasome mediated. In these experiments, 293 cells were transfected with a human immunodeficiency virus (HIV) LTR-driven large Rep-expressing plasmid, either with or without HA-tagged E4orf6. Thirty-six hours later samples were prepared for immunoblotting as described previously (Farris et al., 2010), using anti-Rep78/68 monoclonal 7B73.2 (Hunter & Samulski, 1992) to detect large Rep, and anti-HA antibody (cat# H9658; Sigma) to detect E4orf6. MG132 (10 µM, cat# 474791; Calbiochem) was added to the samples during the last 6 h as indicated. Ad E4orf6 mutants described previously (Nayak & Pintel, 2007), which are deficient in Ad E3 Ub-ligase formation and hence incapable of causing targeted ubiquitination were unable to cause a reduction in accumulated Rep levels (data not shown). Taken together, these results suggest that in addition to a select group of previously reported cellular and viral proteins (Blanchette & Branton, 2009), the Ad E4orf6/E1b 55 kDa E3 Ub-ligase also targets AAV large Rep for proteasomal degradation.
Fig. 1.
AAV large Rep proteins are degraded in the presence of Ad E1b 55 kDa/E4orf6 and are modified by ubiquitination. (a) Immunoblots of samples taken from 293 cells transfected with HIV LTR-driven Rep in the absence (lane 1) or presence of HA-tagged E4orf6 (lanes 2 and 3) as described in the text. Transfections treated with MG132 are shown in lane 3. Tubulin was used as a control for the amount of protein loaded per lane. Rep was detected using anti-Rep78/68 monoclonal 7B73.2 (Hunter & Samulski, 1992), E4orf6 was detected with anti-HA antibody and tubulin with anti-tubulin (cat# T4026; Sigma). (b) Immunoblots of extracts from mock infected (lane 1), AAV alone infected (lane 2), Ad alone infected (lane 3), AAV/Ad infected (lane 4) or AAV/Ad infected with MG132 (10 µM) added, immunoprecipitated with anti Rep 7B73.2 and subsequently immunoblotted with anti-ubiquitin antibody. (c) Immunoblots of extracts taken 48 h following transfection of 293 cells with HIV LTR-driven AAV2 Rep (left panel), CMV-driven AAV5 Rep (right panel), Flag-tagged ubiquitin and HA-tagged E4orf6, as indicated and described in the text, either in the presence or absence of MG132 as shown. Ubiquitinated Rep was detected with anti-Flag antibody (cat# F3165; Sigma). (d) Immunoblots of extracts taken 48 h following transfection of 293T cells with HIV LTR-driven AAV2 Rep, a plasmid expressing HA-tagged ubiquitin allowing extension only on K48 (left panel), a plasmid expressing HA-tagged ubiquitin allowing extension only on K63 (right panel), and E4orf6 as indicated and described in the text, either in the presence or absence of MG132 as shown. Ubiquitinated Rep was detected with anti-HA antibody.
If Rep is a target of the Ad E3 Ub-ligase, we should be able to demonstrate ubiquitination of Rep during AAV/Ad co-infection. As can be seen in Fig. 1(b), during AAV2/Ad5 co-infection, immunoblots of large Rep immunoprecipitated with the anti-Rep78/68 monoclonal 7B73.2 (Hunter & Samulski, 1992) and probed with an anti-ubiquitin antibody (cat # SC-8017; Santa Cruz Biotechnology) revealed high molecular mass ubiquitin conjugates of Rep both in the presence and absence of MG132 (Fig. 1b, lanes 4 and 5). Extracts from mock-infected cells, or cells infected with either AAV or Ad alone showed no specific ubiquitination reactivity (Fig. 1b, lanes 1–3). These experiments were done under stringent washing conditions that we have previously shown fully dissociate cellular proteins, including any potentially Rep-associated, ubiquitinated proteins from our immunoprecipitates (Farris et al., 2010).
To further characterize the ubiquitination of large Rep, we developed a ubiquitination assay utilizing transiently expressed HIV LTR-driven Rep and Flag-tagged ubiquitin. Ubiquitination of both AAV5 and AAV2 large Rep was assayed in 293T cells both in the presence and absence of added E4orf6. In these assays, large Rep was immunoprecipitated using the 7B73.2 antibody, and equivalent amounts of sample were immunoblotted with anti-Flag antibody to detect specific ubiqitination, as described previously (Farris et al., 2010). As expected, both AAV2 (left panel) and AAV5 (right panel) large Reps were ubiquitinated in the presence of E4orf6, which in 293 cells would reconstitute the Ad E3-Ub ligase (Fig. 1c, left panel lane 4 and right panel lane 3), but surprisingly, significant ubiquitination of Rep was also detected in the absence of E4orf6 (Fig. 1c, left panel lane 3 and right panel lane 2). The addition of MG132 had little effect in these experiments (Fig. 1c, left and right panel, lane 5), probably because the immunoprecipitations were subquantitative (data not shown).
There are multiple types of polyubiquitination. Polyubiquitination that extends through lysine 48 of ubiquitin is most often associated with proteasomal degradation (Ciechanover, 1998; Pickart, 1997). The other major form of ubiqutination, extending ubiquitin lysine 63, is most often associated with other modifications of protein function (Komander, 2009). We have previously reported that AAV5 Rep52 is ubiquitinated using both K48 and K63 polyubiquitination (Farris et al., 2010). One possibility that might explain both E4orf6-dependent and E4orf6-independent ubiquitination of large Rep would be that different forms of ubiquitin were added under these different conditions. To further characterize the ubiquitination of large Rep, we used transient expression assays, similar to those described above, and as described previously (Farris et al., 2010), in which mutant forms of HA-tagged ubiquitin that allowed extension only via K48, or alternatively only via K63 (Conze et al., 2008; Lim et al., 2005, 2006; Olzmann et al., 2007), were supplied in trans.
Fig. 1(d) demonstrates that large Rep undergoes both the K48-extension (left panel) and K63-extension (right panel) form of polyubqiuitination. However, surprisingly, both types of ubiquitination were found both in the presence of Ad E4orf6 (Fig. 1d, left and right panel, lanes 4 and 5) and in its absence (Fig. 1d, left and right panels, lane 3), suggesting that the targeting of large Rep for both K48 and K63 ubiquitination can be accomplished by factors other than the Ad E3 Ub-ligase. Ubiquitination patterns were not detected in cell extracts from transfections lacking either Rep or HA-ubiquitin (Fig. 1d, left and right panels, lanes 1 and 2), or in which an HA-tagged ubiquitin that allowed extension only on K29 was included (data not shown).
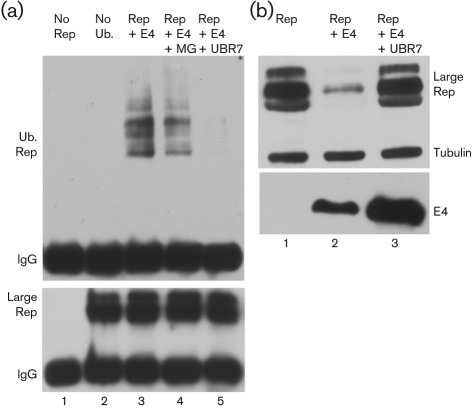
Although we have demonstrated that large Rep can be ubiquitinated via K48 and K63 extension both in the presence and absence of Ad E4orf6, ubiquitination by the Ad E3 Ub-ligase must be an important feature of Ad/AAV interaction because, as shown in Fig. 1(a), ubiquitin-targeted degradation of large Rep is dependent on the E4orf6/E1b 55 kDa Ad E3 Ub-ligase. To demonstrate this involvement more directly, we examined the E4orf6-dependent ubiquitination and targeted degradation of Rep in the presence of the dominant-negative ubiquitin, UBR7 (Sheaff et al., 2000), as described previously (Nayak et al., 2008). As can be seen in Fig. 2, both the E4orf6-dependent ubiquitination (Fig. 2a, compare lanes 3 and 4 to lane 5), and the targeting of large Rep for degradation (Fig. 2b, compare lane 2 to lane 3), was suppressed by the addition of a dominant-negative ubiquitin, implicating the Ad E3-Ub-ligase in this process. For experiments shown in Fig. 2(a), 48 h post-transfection of 293T cells HIV LTR-driven Rep were immunoprecipitated with antibody 7B73.2, and immunoblotted with either anti-Flag (Fig. 2a, top panel) or anti-7B73.2 Rep (Fig. 2a, bottom panel) to detect Flag-tagged ubiquitin or large Rep, respectively. For experiments shown in Fig. 2(b), 293 cells were transfected with HIV LTR-driven Rep either alone, or together with E4orf6 or E4orf 6 plus UBR7, as indicated. Expression of HA-tagged E4orf6 was confirmed with an anti-HA antibody (Fig. 2b, bottom panel). In Fig. 2(a), substantial levels of Rep remain even in the presence of E4orf6 (lane 3 bottom panel) because these immunoprecipitations are subquantitative (data not shown). We have previously shown that the Ad E3Ub-ligase targeted loss of AAV Rep52 evident by Western blotting was not apparent following subquantitative immunoprecipitation of Rep (Farris et al., 2010).
Fig. 2.
E4orf6-dependent ubiqutination and degradation of AAV large Rep is suppressed by the addition of a dominant-negative ubiquitin. (a) Immunoblots of large Rep immunoprecipitated with 7B73.2 and immunoblotted with anti-Flag (top panel) or anti-Rep (7B73.2) (bottom panel) in the presence or absence of the dominant-negative UBR7 or MG132 as indicated, and as described in the text. (b) Immunoblot of samples taken from 293 cells transfected with HIV LTR-driven large Rep alone (lane 1), or with co-expression of E4orf6 (lane 2) or E4orf6 together with UBR7 (lane 3). Large Rep was detected with 7B73.2 and E4orf6 expression was detected by HA (lower panel). Cellular tubulin was used as a loading control.
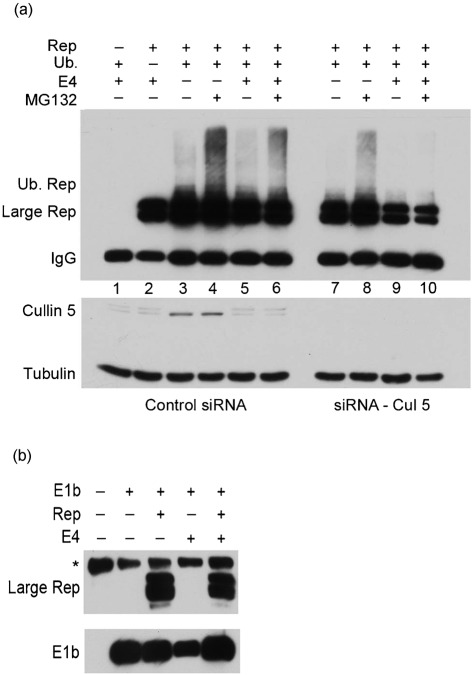
Consistent with the involvement of the Ad E3-Ub-ligase in these processes, siRNA-mediated knockdown [performed exactly as described previously (Farris et al., 2010)] of cullin 5, the cullin present in the Ad E3-Ub-ligase, dramatically reduced E4orf6-dependent (and to a much lesser extent E4orf6-independent) K48 ubiquitination of large Rep (Fig. 3a, compare lanes 9 and 10 to 5 and 6). For these experiments 293T cells were first transfected with either siRNAs to cullin 5 (cat# GS8065; Qiagen) or scrambled siRNAs serving as negative controls. Twenty-four hours following siRNA delivery constructs expressing HIV LTR-driven large Rep, HA-tagged K48 ubiquitin and E4orf6 were transiently transfected. MG132 or DMSO vehicle controls were added in the last 6 h of the experiment. Thirty-six hours later cell lysates were immunoprecipitated with anti-7B73.2 large Rep antibody followed by immunoblotting with an anti-HA antibody to detect specific K48 ubiquitination of large Rep. Specific knockdown of cullin 5 was assessed by Western blotting of equivalent amounts of pre-immunoprecipitation samples.
Fig. 3.
The Ad E3 Ub-ligase participates in the ubiquitination and degradation of AAV large Rep. (a) Top panel: immunoblots of extracts taken 36 h following transfection of 293T cells with HIV LTR-driven AAV2 Rep, an HA-tagged ubiquitin allowing extension only on K48, and E4orf6 as indicated and described in the text, either in the presence or absence of MG132 as shown. In addition, either siRNAs directed toward cullin 5 (Cul 5, lanes 7–10), or scrambled control siRNAs (lanes 1–6) were added as described in the text. Ubiquitinated Rep was detected with anti-HA antibody. Bottom panel: pre-IP Western blot of the same samples in the top panel probed with anti-cullin 5 antibody to monitor specific knockdown of cullin 5. Cellular tubulin was detected as a marker for the amount of protein present in each sample. (b) Immunoblot of cell lysates transfected with plasmids expressing Flag-tagged E1b 55 kDa, large Rep or E4orf6 as indicated. Following transfection, lysates were immunoprecipitated under non-denaturing conditions with an anti-E1b 55 kDa antibody, followed by Western blotting with an anti-large Rep antibody 7B73.2. The lower panel shows the same samples probed with an anti-Flag antibody to demonstrate the presence of E1b 55 kDa. A non-specific band was observed in all lanes as denoted by an asterisk.
Additionally, in the presence of control siRNA, the activated form of cullin 5 [apparent as the higher migrating species of cullin 5 (Chew & Hagen, 2007)] was seen only in the presence of E4orf6 (Fig. 3a, compare lanes 1, 2, 5 and 6 with lanes 3 and 4). This is consistent with cullin 5 participating in the Ad E3 Ub-ligase-targeted ubiquitination of Rep, but not in the ubiquitination of Rep in the absence of this ligase described above. Similar to that observed in Fig. 2(a), significant levels of Rep remain in the presence of E4orf6 (Fig. 3a, lanes 2, 5 and 6), probably because these immunoprecipitations are performed under subsaturating antibody conditions and thus do not reveal quantitative differences (Farris et al., 2010).
We next sought to begin to identify components that could comprise the complex(es) that direct the ubiquitination of large Rep. As can be seen in Fig. 3(b), immunoprecipitation with an anti-E1b 55 kDa antibody (gift of A. J. Berk, UCLA), followed by immunoblotting using the 7B73.2 anti-large Rep antibody, demonstrated that in 293 cells large Rep and E1b 55 kDa form a stable complex both in the presence and absence of E4orf6. Extracts were prepared under non-denaturing conditions and processed as described previously (Farris et al., 2010) with one modification: interfering IgG chains were stripped from the immunoprecipitation elution using the ExactaCruz reagent (cat# SC-45042; Santa Cruz Biotechnology) in order to visualize the E1b 55 kDa protein. This interaction has been reported previously by others as well as ourselves (Nash et al., 2009; Nayak et al., 2008). These results suggest a role for this interaction other than as part of the Ad E3 Ub-ligase and further suggest that within the Ad E3 Ub-ligase complex the E1b 55 kDa protein may interact directly with large Rep irrespective of the presence of E4orf6.
Although the full nature of the roles that the individual helper functions play during AAV infection is not yet fully understood, the Ad E3 Ub-ligase-targeted ubiquitination and proteasomal degradation of AAV large Rep proteins may represent important events in the AAV life cycle.
Acknowledgements
The authors thank Lisa Burger for excellent technical assistance. This work was supported by NIAID with PHS grants R01 046458 and R01 091588.
References
- Berns K. I., Giraud C. (1996). Biology of adeno-associated virus. Curr Top Microbiol Immunol 218, 1–23 [DOI] [PubMed] [Google Scholar]
- Blanchette P., Branton P. E. (2009). Manipulation of the ubiquitin-proteasome pathway by small DNA tumor viruses. Virology 384, 317–323 10.1016/j.virol.2008.10.005 [DOI] [PubMed] [Google Scholar]
- Bowles D., Rabinowitz J. E., Samulski R. J. (2006). The genus Dependoviruses. In Parvoviruses, pp. 15–24 Edited by Kerr J. R., Cotmore S. F. London, UK: Hodder Arnold [Google Scholar]
- Chew E. H., Hagen T. (2007). Substrate-mediated regulation of cullin neddylation. J Biol Chem 282, 17032–17040 10.1074/jbc.M701153200 [DOI] [PubMed] [Google Scholar]
- Ciechanover A. (1998). The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J 17, 7151–7160 10.1093/emboj/17.24.7151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conze D. B., Wu C. J., Thomas J. A., Landstrom A., Ashwell J. D. (2008). Lys63-linked polyubiquitination of IRAK-1 is required for interleukin-1 receptor- and toll-like receptor-mediated NF-κB activation. Mol Cell Biol 28, 3538–3547 10.1128/MCB.02098-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris K. D., Fasina O., Sukhu L., Li L., Pintel D. J. (2010). Adeno-associated virus small rep proteins are modified with at least two types of polyubiquitination. J Virol 84, 1206–1211 10.1128/JVI.01660-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter L. A., Samulski R. J. (1992). Colocalization of adeno-associated virus rep and capsid proteins in the nuclei of infected cells. J Virol 66, 317–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D. (2009). The emerging complexity of protein ubiquitination. Biochem Soc Trans 37, 937–953 10.1042/BST0370937 [DOI] [PubMed] [Google Scholar]
- Lim K. L., Chew K. C., Tan J. M., Wang C., Chung K. K., Zhang Y., Tanaka Y., Smith W., Engelender S., et al. & other authors (2005). Parkin mediates nonclassical, proteasomal-independent ubiquitination of synphilin-1: implications for Lewy body formation. J Neurosci 25, 2002–2009 10.1523/JNEUROSCI.4474-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K. L., Dawson V. L., Dawson T. M. (2006). Parkin-mediated lysine 63-linked polyubiquitination: a link to protein inclusions formation in Parkinson’s and other conformational diseases? Neurobiol Aging 27, 524–529 10.1016/j.neurobiolaging.2005.07.023 [DOI] [PubMed] [Google Scholar]
- Nash K., Chen W., Salganik M., Muzyczka N. (2009). Identification of cellular proteins that interact with the adeno-associated virus rep protein. J Virol 83, 454–469 10.1128/JVI.01939-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak R., Pintel D. J. (2007). Positive and negative effects of adenovirus type 5 helper functions on adeno-associated virus type 5 (AAV5) protein accumulation govern AAV5 virus production. J Virol 81, 2205–2212 10.1128/JVI.02312-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak R., Farris K. D., Pintel D. J. (2008). E4Orf6-E1B-55k-dependent degradation of de novo-generated adeno-associated virus type 5 Rep52 and capsid proteins employs a cullin 5-containing E3 ligase complex. J Virol 82, 3803–3808 10.1128/JVI.02532-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzmann J. A., Li L., Chudaev M. V., Chen J., Perez F. A., Palmiter R. D., Chin L. S. (2007). Parkin-mediated K63-linked polyubiquitination targets misfolded DJ-1 to aggresomes via binding to HDAC6. J Cell Biol 178, 1025–1038 10.1083/jcb.200611128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart C. M. (1997). Targeting of substrates to the 26S proteasome. FASEB J 11, 1055–1066 [DOI] [PubMed] [Google Scholar]
- Sheaff R. J., Singer J. D., Swanger J., Smitherman M., Roberts J. M., Clurman B. E. (2000). Proteasomal turnover of p21Cip1 does not require p21Cip1 ubiquitination. Mol Cell 5, 403–410 10.1016/S1097-2765(00)80435-9 [DOI] [PubMed] [Google Scholar]
- Weger S., Hammer E., Heilbronn R. (2004). SUMO-1 modification regulates the protein stability of the large regulatory protein Rep78 of adeno associated virus type 2 (AAV-2). Virology 330, 284–294 10.1016/j.virol.2004.09.028 [DOI] [PubMed] [Google Scholar]