Abstract
The hallmark attribute of North American West Nile virus (WNV) strains has been high pathogenicity in certain bird species. Surprisingly, this avian virulent WNV phenotype has not been observed during its geographical expansion into the Caribbean, Central America and South America. One WNV variant (TM171-03-pp1) isolated in Mexico has demonstrated an attenuated phenotype in two widely distributed North American bird species, American crows (AMCRs) and house sparrows (HOSPs). In order to identify genetic determinants associated with attenuated avian replication of the TM171-03-pp1 variant, chimeric viruses between the NY99 and Mexican strains were generated, and their replicative capacity was assessed in cell culture and in AMCR, HOSP and house finch avian hosts. The results demonstrated that mutations in both the pre-membrane (prM-I141T) and envelope (E-S156P) genes mediated the attenuation phenotype of the WNV TM171-03-pp1 variant in a chicken macrophage cell line and in all three avian species assayed. Inclusion of the prM-I141T and E-S156P TM171-03-pp1 mutations in the NY99 backbone was necessary to achieve the avian attenuation level of the Mexican virus. Furthermore, reciprocal incorporation of both prM-T141I and E-P156S substitutions into the Mexican virus genome was necessary to generate a virus that exhibited avian virulence equivalent to the NY99 virus. These structural changes may indicate the presence of new evolutionary pressures exerted on WNV populations circulating in Latin America or may signify a genetic bottleneck that has constrained their epiornitic potential in alternative geographical locations.
Introduction
Since West Nile virus (WNV) was first identified in New York City in 1999, evidence for viral transmission has been documented throughout the western hemisphere, indicating the potential establishment of endemic transmission cycles as far south as Argentina (Adrián Diaz et al., 2008). Surveillance studies conducted in Mexico (Estrada-Franco et al., 2003; Farfán-Ale et al., 2004, 2006), Central America (Morales-Betoulle et al., 2006), the Caribbean (Barrera et al., 2008; Dupuis et al., 2003; Komar et al., 2005) and South America (Bosch et al., 2007; Mattar et al., 2005) have provided serological evidence of transmission of WNV but have generated few WNV isolates, possibly indicating limited or highly focal transmission of the virus within Latin America (Bosch et al., 2007; Dupuis et al., 2005; Komar & Clark, 2006). Avian virulence, a common feature associated with North American WNV transmission in the USA and Canada, has not been reported in Latin America or the Caribbean, and plaque variants of a WNV isolate from Tabasco State, Mexico, in 2003 have demonstrated reduced avian viraemia production and virulence in American crows (AMCRs) and house sparrows (HOSPs) (Brault et al., 2011). The implementation of WNV dead-bird surveillance programmes in North America has enhanced surveillance efforts (Eidson, 2001), serving to foreshadow WNV epizootics, and has provided an invaluable source of viral isolates for genotypic and phenotypic characterization (Guptill et al., 2003; Komar, 2001; Mostashari et al., 2003). The paucity of avian mortality in Latin America has undoubtedly contributed to the dearth of knowledge of enzootic transmission mechanisms in Latin America.
The basis for the apparent lack of avian mortality in Latin America has not been demonstrated conclusively; however, several plausible explanations to account for this observation include: (i) pre-existing immunity in avian populations to closely related flaviviruses, reducing the pathogenesis of secondary WNV infections (Farfán-Ale et al., 2009); (ii) natural resistance of avian species in Latin America to lethal WNV infection (Reed et al., 2009); (iii) dampened transmission associated with increased avian biodiversity in more tropical latitudes (Ezenwa et al., 2006; Johnson & Thieltges, 2010; Swaddle & Calos, 2008); (iv) reduced surveillance efforts; (v) reduction in the avian virulence potential of a founding strain; and (vi) selection for a genotype with reduced avian virulence potential. The isolation of a murine and avian attenuated WNV variant (TM171-03-pp1) from a raven in Tabasco State, Mexico (Beasley et al., 2004), with reduced avian pathogenic properties (Brault et al., 2011) indicates that strains with altered pathogenic characteristics have circulated in Mexico. Similarly attenuated strains have also been identified in Texas (Brault et al., 2011; Davis et al., 2004).
Isolates of WNV from Northern Mexico (Davis et al., 2005; Elizondo-Quiroga et al., 2005) in close proximity to the US border as well as in Puerto Rico (Barrera et al., 2008) have been demonstrated to be most synonymous with the North American genotype (WN02), which, subsequent to 2002, has largely displaced the East Coast North American founding genotype (NY99). The lack of sequence data available on WNV isolates collected in the Caribbean and much of Latin America has precluded the specific identification of genotypic changes associated with transmission in these ecosystems. The Mexican TM171-03 isolate from the brain of a raven consists of mixed-plaque phenotypes: a small-plaque variant (TM171-03-pp1; GenBank no. JN051152) lacking a functional envelope protein (E) glycosylation motif (NYP, at aa 154–156) and a large-plaque variant (TM171-03-pp5; GenBank no. JN051153) exhibiting a functional NYS glycosylation motif (Beasley et al., 2004). In a murine model, E protein N-glycosylation of North American WNV strains has been positively correlated with neuroinvasive capacity and disease severity (Beasley et al., 2005; Shirato et al., 2004). The murine virulence phenotype did not appear to be attenuated by the presence of additional amino acid substitutions [I141T in the pre-membrane protein (prM), I245V in non-structural protein NS4B and T898I in non-structural protein NS5] or the nucleotide substitutions in the 5′- and 3′UTRs of the genome of the glycosylated TM171-03-pp5 variant (Beasley et al., 2004). Avian studies utilizing the non-glycosylated WNV TM171-03-pp1 variant demonstrated a decreased peripheral replication in AMCRs and HOSPs, with higher survival rates observed in the AMCR model when compared with avian virulent NY99 and TWN301 isolates (Brault et al., 2011). The glycosylation-competent plaque-purified variant WNV TM171-03-pp5 also demonstrated an attenuated phenotype in AMCRs and HOSPs compared with the NY99 isolate, indicating the contribution of other amino acid residues or non-coding nucleotide changes to reduced replication of the WNV TM171-03 plaque variants in avian hosts.
In order to assess directly the role of N-linked glycosylation and accessory mutations in the attenuation of avian replication of the TM171-03-pp1 isolate, non-synonymous genetic differences between the WNV TM171-03-pp1 variant and NY99 virus were introduced into a virulent WNV NY99 infectious cDNA clone (WN/IC-P991) (Kinney et al., 2006) singly or in combination (Table 1). The replication phenotypes of viruses generated from these recombinant constructs were tested in vitro in HD11 avian macrophages and in vivo in AMCRs for a direct assessment of the relative attenuating role of TM171-03-pp1 mutations, singly or in combination. To assess whether the attenuating effects of substitutions were avian species specific, viraemias were also assessed in two additional passerine bird species, HOSPs and house finches (HOFIs).
Table 1. Genetic differences between parental (WN/IC-P991 and WN-MX03) and point-mutant viruses generated.
Nucleotide numbers correspond to WNV NY99 (strain 382-99) genomic positions. Nucleotide positions are presented for 5′- and 3′UTR mutations and amino acid positions within respective genes are presented for amino acid changes. Amino acid substitutions are presented in upper case and nucleotide substitutions in lower case. Dashes represent the same nucleotide or amino acid identity compared with the WN/IC-P991 sequence.
| Parental/mutant recombinant virus | 5′UTR (nt 50) | 5′UTR (nt 71) | prM (aa 141) | E (aa 156) | NS4B (aa 245) | NS5 (aa 898) | 3′UTR (nt 10 828) | 3′UTR (nt 10 989) |
| WN/IC-P991 (NY99)* | a | a | I | S | I | T | t | g |
| WN/IC-5′ | g | g | – | – | – | – | – | – |
| WN/IC-prM | – | – | T | – | – | – | – | – |
| WN/IC-E | – | – | – | P | – | – | – | – |
| WN/IC-5′.prM.E | g | g | T | P | – | – | – | – |
| WN/IC-5′.E | g | g | – | P | – | – | – | – |
| WN/IC-prM.E | – | – | T | P | – | – | – | – |
| WN/IC-NS4B | – | – | – | – | V | – | – | – |
| WN/IC-E.NS4B | – | – | – | P | V | – | – | – |
| WN/IC-NS4B.NS5 | – | – | – | – | V | I | – | – |
| WN/IC-3′ | – | – | – | – | – | – | g | a |
| WN/IC-.E.3′ | – | – | – | P | – | – | g | a |
| WN/IC-5′.E.3′ | g | g | – | P | – | – | g | a |
| WN-MX03/IC-prM.E | g | g | – | – | V | I | g | a |
| WN-MX03/IC | g | g | T | P | V | I | g | a |
| TM171-03-pp1† | g | g | T | P | V | I | g | a |
GenBank accession no. AF196835 (Lanciotti et al., 1999). These substitutions represent two additional genetic differences between the WN/IC-P991 and WN-MX03 strains presented within this table.
Nucleotide and amino acid differences between the parental NY99 WNV (strain 382-99; synonymous with the exception of the abovementioned differences with WN/IC-P991) and the TM171-03-pp1 consensus sequence.
Results
Generation and phenotypic characterization of WNV NY99/MX03 mutants
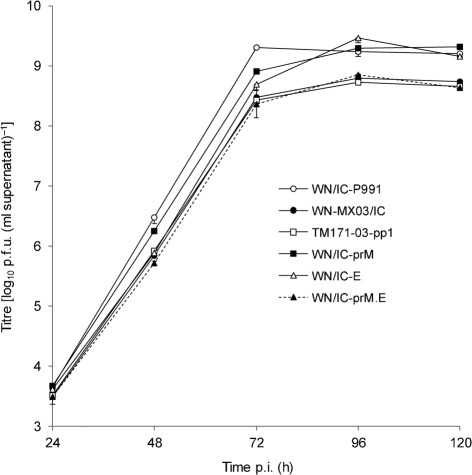
Mutagenesis was performed on both 5′ and 3′ plasmid cassettes, and differential ligation of mutated and parental plasmids was used for the successful generation of 14 recombinant viruses from WNV NY99 and the Mexican TWN171-03 clone-derived virus containing the same prM-141T and E-156P residues (WN-MX03/IC) (Table 1). Infectious virus was rescued for all recombinant constructs and all viruses grew to at least 8 log10 p.f.u. ml−1 by 96 h post-infection (p.i.) (the growth profile of a subset of viruses is shown in Fig. 1). Initial plaque morphologies were visualized in Vero cells using crystal violet to stain the cell monolayer, and viral plaque diameters were measured. At 72 h p.i., the parental WNV TM171-03-pp1 plaque variant exhibited a mean diameter of 2.1±0.6 mm that was indistinguishable from the WN-MX03/IC virus (1.9±0.5 mm) and WN/IC-prM.E virus containing both the prM-141T and E-156P mutations (1.9±0.3 mm) (Table 2). The WN/IC-E mutant with the E-S156P substitution exhibited an intermediate plaque size phenotype of 3.2±0.3 mm, distinguishable from both the WN/IC-P991 and WN-MX03/IC viruses (P<0.05). In contrast, the WN/IC-P991 wild-type virus and mutant WN-MX03/IC-prM.E virus (containing the prM-141I and E-156S mutations in the TM171-03-pp1 backbone) exhibited an indistinguishable large-plaque phenotype in Vero cells (4.5±0.4 and 4.4±0.3 mm, respectively), implicating a role of both the prM and E substitutions in the small-plaque phenotype of the TM171-03-pp1 plaque variant. Small reductions in growth in Vero cells were also identified for WN/IC mutants containing either the prM-I141T (WN/IC-prM ) or E-S156P (WN/IC-E) substitution; however, the complete 11-fold reduction in titre identified with the TM171-03-pp1 variant was achieved only by incorporation of both structural mutations (WN/IC-prM.E; Fig. 1).
Fig. 1.
Mean viral titre±sd of triplicate cultures of parental and NY99/MX03 point-mutant viruses in Vero cells inoculated at an m.o.i of 0.01, with a detection limit of 50 p.f.u. ml−1.
Table 2. Mean plaque diameter of parental and NY99/MX03 WNV chimeric viruses at 72 h p.i.
| Virus | Plaque diameter (mm±sd) |
| WN/IC-P991 | 4.5±0.4 |
| WN-MX03/IC | 1.9±0.5 |
| TM171-03-pp1 | 2.1±0.6 |
| WN/IC-5′.E.3′ | 3.3±0.3 |
| WN/IC-E | 3.2±0.3 |
| WN/IC-prM.E | 1.9±0.3 |
| WN-MX03/IC-prM.E | 4.4±0.3 |
Avian monocyte growth phenotype characterization
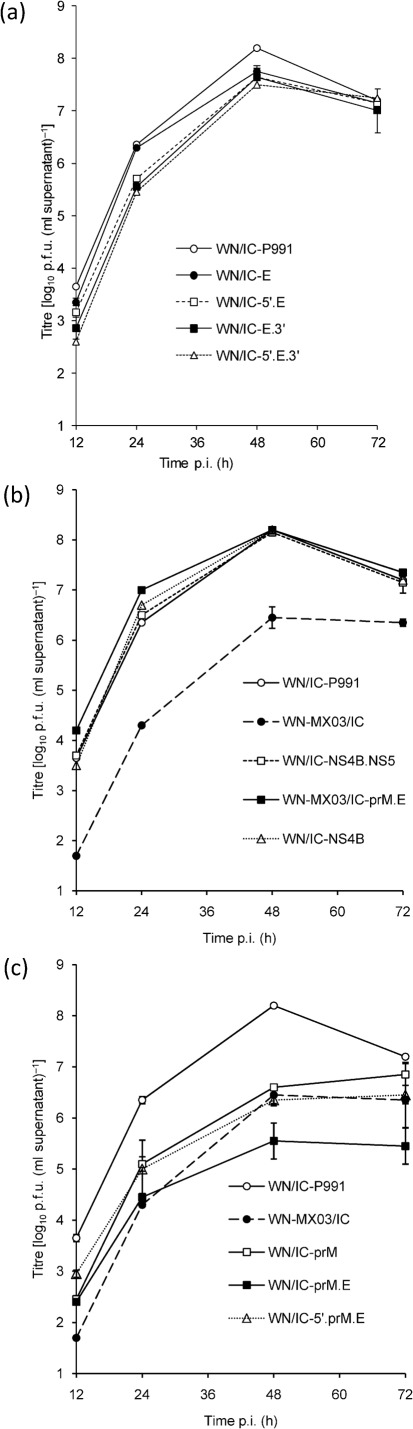
As previous studies have implicated the importance of monocyte-derived cells as sites of initial virus replication and differential induction of innate immune responses (Garcia-Tapia et al., 2007; Silva et al., 2007; Steele et al., 2000; Wünschmann et al., 2004), all WN/IC mutants generated (Table 1) were assessed for growth in an HD11 chicken monocyte line (kindly provided by Dr Kirk Klasing, University of California, CA, USA) to identify potential in vitro markers for avian attenuation of the WNV TWN171-03-pp1 variant. WN/IC mutant viruses containing the E-156P substitution alone (WN/IC-E) or with the TM171-03-pp1 5′- and/or 3′UTR nucleotide changes with or without the E mutation [WN/IC-5′, WN/IC-3′ (not shown in Fig. 1a), WN/IC-5′.E.3′, WN/IC-E.3′ and WN/IC-5′.E) did not demonstrate significantly retarded growth (P>0.05) based on the growth-curve comparisons or peak titres compared with the parental WN/IC-P991 virus (Fig. 1a). Similarly, recombinant viruses containing the TM171-03-pp1 NS4B-245V singly (WN/IC-NS4B) or the NS4B and NS5-898I amino acid substitutions in combination (WN/IC-NS4B.NS5) in the WN/IC backbone also failed to exhibit titres distinguishably lower than the wild-type WN/IC virus (P>0.1; Fig. 2b). Introduction of the prM-141T mutation into the WN/IC genome (WN/IC-prM) significantly lowered (P<0.001) the mean peak virus titre by 40-fold when compared with the WN/IC-P991 virus at 48 h p.i. (Fig. 2c). When both structural TM171-03-pp1 mutations (prM-141T and E-156P) were introduced into the WN/IC backbone in concert (WN/IC-prM.E), the mean peak virus titre was observed to be 450-fold lower than the WN/IC-P991 wild-type virus at 48 h p.i. (P<0.001) (Fig. 2c). However, WN-MX03/IC, the Mexican TWN171-03 clone-derived virus containing the same prM-141T and E-156P residues, exhibited only a 56-fold lower mean peak titre compared with the WN/IC-P991 virus (P<0.01) at 48 h p.i. (Fig. 2c), indicating a potential modulating effect afforded by alternative non-structural genetic changes in the TM171-03-pp1 backbone. The WN/IC-5′.prM.E mutant demonstrated a peak viraemia indistinguishable from that of the WN-MX03/IC virus (Fig. 2c), indicating that a modulating effect was imparted by the two 5′UTR mutations in HD11 cells. Inclusion of the prM-141I and E-156S mutations in the WN-MX03/IC backbone resulted in a virus (WN-MX03/IC-prM.E) that was indistinguishable from the WN/IC-P991 in HD11 cells (Fig. 2b), supporting the dominant attenuating role of the combined prM and E mutations for retarded growth of the MX03 virus in HD11 cells.
Fig. 2.
In vitro growth profiles of parental genotype and NY99/MX03 point-mutant viruses in an avian myeloid cell line (HD11 chicken monocytes). The mean viral titre±sd was determined from triplicate cultures inoculated at an m.o.i of 0.01, with a detection limit of 50 p.f.u. ml−1. The parental recombinant WN/IC-P991 (a–c) and WN-MX03/IC (b, c) were included for reference. All samples were taken from a single concurrent experiment and data were partitioned into separate graphs for presentation purposes.
TM171-03-pp1 genetic determinants of attenuated avian replication
The viraemia in AMCRs following inoculation with virus derived from the WN-MX03/IC infectious cDNA was indistinguishable from that observed from the WNV TM171-03-pp1 variant (Fig. 3a), indicating the utility of this clone as a surrogate backbone for the incorporation of NY99-restoring avian virulence mutations. The mean peak viral load for AMCRs inoculated with the WN/IC-E virus was observed at 5 days p.i. compared with 4 days p.i. for the parental WN/IC-derived virus and was 1000-fold lower (P<0.006) (Fig. 3b). This attenuated phenotype was observed following inoculation of AMCRs with all WNV NY99/MX03 mutants containing the E-S156P substitution (Fig. 3b and Supplementary Fig. S1, available in JGV Online). Inclusion of the NS4B and UTR genetic elements in various combinations with the E mutation failed to significantly modulate AMCR viraemia compared with the WN/IC-E virus (Supplementary Fig. S1). Both the wild-type WN/IC-P991 and WN/IC-prM mutant manifested mean peak viral titres at 4 days p.i., with the WN/IC-prM virus producing a threefold lower viraemia at that time point (P>0.4; Fig. 3a). Peak titres observed for infection with the WN/IC-prM (Fig. 3a) and WN/IC-E (Fig. 3b) viruses were significantly different (P<0.05). Mortality appeared to correlate well with the viraemia profiles observed in AMCRs (Fig. 4). Inoculation of the TM171-03-pp1 virus resulted in only 50 % mortality, and the clone-derived WN-MX03/IC virus (n = 3; not shown in Fig. 4) elicited no mortality, whereas the WN/IC-P991 virus yielded 100 % mortality (n = 8) within 6 days (Brault et al., 2011). A lower-than-expected mortality for the WN-MX03/IC virus compared with the parental TM171-03-pp1 virus was attributed to the small sample size, as viraemias generated by these two viruses were not distinguishable in AMCRs or HOSPs/HOFIs (Figs 4 and 5). Incorporation of the MX03 prM mutation into the WN/IC virus (WN/IC-prM) had a minimal effect on virulence, with only a single survivor being observed out of the six inoculated AMCRs (Fig. 4). Two of eight (25 %) AMCRs inoculated with the recombinant WNV containing the MX03 E mutation (WN/IC-E) survived. The recombinant virus containing both the prM and E MX03 mutations (WN/IC-prM.E) demonstrated a mortality rate of 50 % (n = 6), identical to that of the parental TM171-03-pp1 strain (Fig. 4).
Fig. 3.
(a, b) Selected viraemia profiles of American crows inoculated with 1500 p.f.u. parental genotype and NY99/MX03 point-mutant viruses. Crows were inoculated with WN/IC-P991 (n = 8), WN/IC-E (n = 8), TM171-03-pp1 (n = 8), WN/IC-prM (n = 6), WN/IC-prM.E (n = 6) and WN-MX03/IC (n = 3). The TM171-03-pp1 viraemia profile was taken from previously published data (Brault et al., 2011). Titres were determined by standard plaque assay in Vero cells with a limit of detection of 1.7 log10 p.f.u. (ml serum)−1. Results are shown as means±sd, and only positive or negative values are presented for clarity. The WN/IC-P991 and WN-MX03 viraemia profiles have been plotted on both graphs for reference. Viraemia profiles of additional chimeras have been omitted for clarity of presentation and are available as a Supplementary Fig. S1 (available in JGV Online).
Fig. 4.
Percentage survival of AMCRs inoculated with 1500 p.f.u. parental and NY99/MX03 point-mutant viruses and serially bled, as presented in Fig. 3 (n = 8 for WN/IC-P991, TM171-03-pp1 and WN/IC-E; n = 6 for WN/IC-prM and WN/IC-prM.E). The TM171-03-pp1 mortality plot has been reproduced for reference from previously published data (Brault et al., 2011).
Fig. 5.
(a, b) Viraemia profiles of HOSPs (n = 6; except for WN/IC-P991 where n = 5) following inoculation with 1500 p.f.u. NY99/MX03 parental genotype and point-mutant viruses. Daily mean peak titres are represented (±sd) with a detection limit of 1.7 log10 p.f.u. (ml serum)−1. WN/IC-P991 and WN-MX03/IC have been displayed on both graphs for comparative purposes.
The attenuating growth capacity of the WNV TM171-03-pp1 structural mutations (prM-141T and E-156P) was also assessed in HOSPs, a common avian species that has been implicated previously as an important reservoir host for WNV (Reisen et al., 2005). Unlike the viraemia data generated for AMCRs, significant variability in the mean titres observed in HOSPs within viral infection groups confounded statistical analyses. This variability resulted from the failure of some birds within the groups to generate viraemia, despite seroconversion at 21 days p.i. As such, the limit of detection titre [1.7 log10 p.f.u. (ml−1 serum)] was used for calculation of means. The WN/IC-prM and WN/IC-E viruses had one HOSP each and the WN/IC-prM.E infection group had two HOSPs that were seropositive, yet failed to generate detectable viraemias. The E-156P mutation reduced the magnitude of viraemia compared with the WN/IC-P991 virus in HOSPs by 91-fold at 3 days p.i. (Fig. 5a); however, the large variability in titres at this point for the prM mutant [range 1.7–6.4 log10 p.f.u. (ml serum)−1] obscured any inference of statistical significance (P>0.8) between the prM and E mutants. However, peak viraemias in HOSPs were identified at 3 days p.i. following infection with the WN/IC-prM mutant virus, with this mutant exhibiting a mean peak viral load 334-fold lower than the WN/IC-P991 virus (P>0.4) at this time point (Fig. 5a). When comparing the two TM171-03-pp1 structural mutations in the HOSP model, the WN/IC-prM mutant virus produced only an approximately fourfold lower mean peak viraemia than WN/IC-E (P>0.5) (Fig. 5a). Inclusion of both WNV TM171-03-pp1 structural mutations (WN/IC-prM.E) drastically reduced the mean peak viraemia by 57 000-fold when compared with the WN/IC-P991 virus (P<0.001) (Fig. 5b). Reciprocally, incorporation of the prM-141I and E-156S mutations into the TM171-03-pp1 backbone (WN-MX03/IC-prM.E) restored the high-viraemia phenotype in HOSPs (Fig. 5b), which was indistinguishable from the WN/IC-P991 parental virus [6.4 vs 7.2 log10 p.f.u. (ml serum)−1; P>0.5].
The cumulative attenuating effect of the prM-I141T and E-S156P substitutions on viraemia was also assessed in HOFIs. Similar to the results observed in HOSPs, HOFIs inoculated with the WN/IC-prM.E mutant developed a mean peak viraemia that was 38 000-fold lower than the parental WN/IC-P991 virus (P<0.001) and was indistinguishable from the WN-MX03/IC virus (P>0.1) (Fig. 6). Restoration of the high-viraemia phenotype was observed in HOFIs inoculated with the mutant virus in which the prM-141I and E-156S substitutions had been incorporated into the TM171-03-pp1 clone backbone (WN-MX03/IC-prM.E). Peak viraemias in HOFIs inoculated with this mutant reached 7.4 log10 p.f.u. (ml serum)−1 and were indistinguishable from the WN/IC-P991 virus (P>0.5).
Fig. 6.
Viraemia profiles of HOFIs (n = 6) following inoculation with 1500 p.f.u. parental genotype and NY99/MX03 point-mutant WNV recombinant viruses. Daily mean peak titres are represented (±sd) with a detection limit of 1.7 log10 p.f.u. (ml serum)−1.
Sequence analyses
All desired nucleotide/amino acid changes were present and no additional non-synonymous amino acid substitutions or non-coding nucleotide changes were introduced while generating the 14 chimeric WN/IC NY99/MX03 viruses (Table 1). Amplicons generated from peak viraemia serum sample time points of a select number of AMCRs inoculated with the WN/IC-prM.E virus were sequenced and compared with the consensus sequence from the viral inoculum. An amplicon of ~2 kb consisting of a portion of the WNV prM and E genes (genomic positions 301–2206) was sequenced. Sequencing of amplicons from all AMCR sera failed to identify any mutations at the prM-141 and/or E-156 amino acid positions or alternative compensatory mutations within this gene region. Additionally, sequence analyses performed on viral RNA extracted from the brains of AMCRs that succumbed to infection demonstrated no reversions or compensatory amino acid changes in these same gene regions.
Discussion
The epidemiological and/or biological basis for the paucity of WNV isolations from Central/South America, Mexico and the Caribbean has not been explored fully. The recent incursion into North America by WNV has coincided with a reduction in St Louis encephalitis virus (SLEV) activity in certain enzootic habitats within the USA, suggesting that WNV could potentially displace enzootic transmission by flaviviruses imparting heterologous immunity in similar maintenance hosts (Gibbs et al., 2006; Reisen et al., 2008). These epidemiological findings have been supported by experimental studies that indicate a higher replicative fitness of WNV in avian studies compared with SLEV genotypes (Reisen et al., 2005), as well as a reduced susceptibility to infection of previously flavivirus-infected mosquitoes (Pesko & Mores, 2009). Serological cross-protection between these flaviviruses in HOFIs has been demonstrated to result in a diminished avian viraemia following heterologous flavivirus challenge, providing further evidence to support cross-neutralizing flavivirus immunity as a potential factor dampening the force of transmission in more tropical latitudes where transmission of a more expansive group of flaviviruses has been described (Fang & Reisen, 2006). Competition with other closely related flaviviruses such as Bussuquara, Ilheus and Rocio viruses and SLEV for naïve vertebrate hosts could reduce the force of transmission by dampening avian viraemia and diminishing WNV-associated avian mortality observed in Latin America and the Caribbean (Gubler, 2007). Monitoring WNV consensus genotypes within emerging transmission foci for genetic changes influenced by selective constraints imposed by transmission within new ecosystems will be critical for assessing viral phenotypic modulation in response to adaptation to new ecological niches.
Since the first documentation of WNV incursion into Latin America in 2003, a scarcity of isolates has been reported, despite widespread serological evidence of viral transmission (Johnson et al., 2010). Most isolates have been found in Mexico in close proximity to the US border and in Puerto Rico (Barrera et al., 2008; Deardorff et al., 2006); interestingly, these viruses have been representative of the North American genotype and have shared high degrees of genetic identity with viruses circulating in the USA. In contrast, the first WNV isolate (TM171-03) collected beyond the USA/Mexican border was isolated from a raven in Tabasco State in 2003. Phylogenetic analysis indicated this virus to be a member of the East Coast genotype, and introduction of this virus probably occurred via bird migration from the south-eastern USA directly or by way of the Caribbean (Deardorff et al., 2006). Reduced neuroinvasiveness in mice (Beasley et al., 2004) has been genetically correlated with the deletion of a single glycosylation motif within the E protein (E-S156P) that limited peripheral virus replication and dissemination into secondary tissues including the brain. The consensus sequence of the TM171-03-pp1 isolate identified prM-I141T, E-S156P, NS4B-I245V and NS5-T898I amino acid substitutions and three 5′UTR and five 3′UTR nucleotide changes compared with the WNV NY99 isolate (Beasley et al., 2004). A glycosylated large-plaque-forming variant (TM171-03-pp5) and a non-glycosylated E protein small-plaque mutant (TM171-03-pp1) of the WNV TM171-03 isolate demonstrated different degrees of attenuation in AMCRs and HOSPs when compared with NY99 (Brault et al., 2011), with the non-glycosylated plaque variant displaying greater attenuation in both avian species than the glycosylated virus. However, in addition to having a functional glycosylation motif, the large-plaque variant (TM171-03-pp5) also contained a NS5-R224G amino acid substitution and one 5′UTR and one 3′UTR nucleotide difference (mixed in TM171-03-pp1) compared with the small-plaque variant (TM171-03-pp1) (Beasley et al., 2004), thus requiring the use a reverse genetics approach to conclusively implicate the genetic determinant(s) associated with altered avian fitness of these plaque variants. This approach also allowed the introduction of various amino acid substitution combinations to identify synergistic and/or compounding effects of multiple amino acids on the viral phenotype in birds. Incorporation of the prM-I141T or E-S156P mutations individually resulted in viruses exhibiting varying degrees of growth attenuation that was avian species specific. A virus mutant containing the E-S156P substitution, eliminating the N-linked glycosylation motif, exhibited significantly altered peripheral growth in the highly susceptible AMCRs but had only a modest effect in HOSPs. Incorporation of the prM-I141T substitution into the WNV NY99 backbone alone significantly lowered viraemia in HOSPs but alone had no discernible effect in AMCRs. Introduction of both prM-I141T and E-S156P substitutions into the NY99 genetic backbone was required to fully attenuate the virus to the level of the TM171-03-pp1 plaque variant in AMCRs and HOSPs. Single mutants were not assessed in HOFIs; however, inoculation of HOFIs with the double mutant demonstrated a viraemia profile indistinguishable from that of the TM171-03-pp1 plaque variant. Interestingly, restoration of virulence levels indistinguishable from the parental NY99 strain was identified when both HOSPs and HOFIs were inoculated with the mutant virus containing both prM-141I and E-156S substitutions in the TM171-03-pp1 genetic backbone.
Together, these structurally non-conserved amino acid changes could impact host immune recognition and/or receptor-binding efficiency, as the prM and E proteins interact closely with each other to shape the outer-virion surface composition. On the surface of immature or partially mature WNV virions, the prM and E proteins interact to form stable trimers in which the prM restricts premature rearrangement of the E protein and subsequent fusion in low-pH secretory vesicles prior to exocytosis from the cell (Stiasny & Heinz, 2006). The fusion loop on domain II of the WNV E protein is protected until ~90 aa of the prM are cleaved by a cellular furin protease during exportation through the secretory pathway in the host Golgi. The subsequent cleavage of prM to M protein results in a structural rearrangement of the envelope homodimers for presentation of the receptor-binding motifs in domain III, resulting in the formation of a mature infectious virion (Heinz & Allison, 2000). The prM-I141T mutation is predicted to lie within a transmembrane domain involved in anchoring the prM protein to the lipid membrane of the host-cell endoplasmic reticulum (Li et al., 2008). WNV prM amino acid alignments demonstrate this transmembrane domain to be highly conserved, and the isoleucine at position 141 has been found in all other WNV isolates analysed (Misra & Schein, 2007). Analysis of dengue virus prM–E crystal structures has revealed that large conformational changes probably occur in the prM protein transmembrane regions during low-pH-induced virion maturation to infectious viral particles (Zhang et al., 2003). The less hydrophobic prM-I141T amino acid substitution could potentially hinder the low-pH conformational modifications of the prM protein that expose the viral furin cleavage site to cellular furin. This could directly impact on the proportion of immature envelope protein trimers versus mature dimers presented on the virion surface.
Previous studies have demonstrated that virus maturation impacts host neutralizing antibody recognition of viral epitopes, and the percentage of mature dimers on the virion surface is a major determinant of viral receptor-mediated infectivity (Heinz et al., 1994; Nelson et al., 2008). Decreased furin cleavage efficiency of the prM and E protein instability, due to a lack of N-linked glycans, could contribute to the attenuated phenotype observed in this study. Incorporation of the prM and E mutations individually demonstrated some level of reduced viral growth within the avian in vitro and in vivo systems reported here; however, it is unclear whether the attenuation effects of these mutations are cumulative or synergistic. The presence of mutations identified in these studies that restrict viral growth in avian hosts could be the result of immune selective pressures imposed by pre-existing flaviviral immunity in avian hosts. Future studies are warranted to assess the relative fitness of mutant viruses expressing these genetic changes compared with parental NY99 and TM171-03-pp1 viruses in the presence of avian immunoglobulin generated to heterologous flaviviral infections. Alternatively, the presence of highly attenuating mutations in circulating WNV strains in Mexico could have resulted from genetic drift in which the introduction of a limited viral genetic population into Latin America, perhaps by a migratory bird, as has been suggested previously (Deardorff et al., 2006), could have randomly initiated transmission with a low-fitness variant. Once available, analyses of more WNV sequences from transmission foci in Mexico, Central America, the Caribbean and South America will allow further assessment of this founder-effect theory. Regardless of the evolutionary forces that have resulted in a genotype of WNV isolated from Mexico with reduced avian viraemia potential, it is apparent that, despite the reduced growth capacity in avian hosts reported here, these viruses maintain the potential to be transmitted by certain highly susceptible avian hosts. Viruses that contained both prM and E mutations generated peripheral titres as high as 5 log10 p.f.u. (ml serum)−1 in AMCRs and HOFIs, a level likely to be permissive for infection of competent Culex spp. mosquito vectors (Dohm & Turell, 2001; Dohm et al., 2002). The significantly diminished peripheral viraemias in birds would undoubtedly reduce the efficiency of infection of enzootic mosquito vectors and diminish the force of transmission where these genotypes circulate. Such a reduction in the force of transmission is consistent with the low incidence of human and equine disease reported in parts of Latin America. Furthermore, the reduced viral growth due to the presence of these mutations could also explain the lack of avian mortality observed south of the USA–Mexico border. Additional studies utilizing sympatric avian hosts with locally isolated WNV isolates will be needed to address this hypothesis further.
Methods
Plasmids and infectious cDNA clones.
All WNV TM171-03-pp1/NY99 point mutants and chimeras were generated using a two-plasmid infectious clone system (pWN-AB and pWN-CG) described previously (Kinney et al., 2006). Site-directed mutagenesis was carried out on pWN-AB cDNA to introduce single or combinations of TM171-03-pp1 structural amino acid (prM-I141T and E-S156P) and 5′UTR mutations. Similarly, mutagenesis was performed for incorporation of individual or combinations of mutations of the NS4B-I245V, NS5-T898I and 3′UTR mutations, with pWN-CG. Different pairings of mutated and wild-type pWN-AB and pWN-CG cDNAs were ligated in vitro, as described previously (Kinney et al., 2006), to generate the various mutations. Transcription of the resulting full-length cDNAs was performed using a T7 RNA AmpliScribe kit with 6 mM m7G (5′)ppp-(5′)A cap analogue based on the manufacturer’s instructions (Epicentre). In vitro-transcribed RNA was transfected into baby hamster kidney cells using a Petri pulser (BTX), propagated once in Vero cells and quantified by plaque assay in Vero cells for use in all in vitro and in vivo experiments. Viral RNA was extracted from the viral stocks, and the complete genomes of all rescued viruses were sequenced as described previously (Kinney et al., 2006) to verify maintenance of the desired mutations in all rescued viruses.
In vitro growth profiles.
Select WN/IC mutants were inoculated onto Vero and avian macrophage HD11 cells at an m.o.i. of 0.01 in triplicate using six-well plates. Briefly, the viral inoculum was allowed to adsorb for 1 h at 37 or 41 °C for Vero and HD11 cells, respectively. Monolayers were washed twice with PBS, and 4.5 ml Dulbecco’s minimal essential medium (DMEM; Vero cells) and RPMI 1640 (HD11 cells) supplemented with 5 % FBS and 100 U penicillin/streptomycin ml−1 was added to each well. One hundred microlitres of supernatant was collected at 12, 24, 48 and 72 h p.i. for HD11 cells and every 24 h p.i. for 5 days for Vero cultures. Sampled supernatants were diluted 1 : 10 using the same culture medium containing 20 % FBS and stored at −80 °C until titration by standard plaque assay in Vero cells, as described previously (Brault et al., 2004). This dilution and the plating of a 200 µl inoculum for plaque assays resulted in a detection limit of 50 p.f.u. (ml supernatant)−1.
Avian infection and sero-sampling.
Wild-caught AMCRs, HOSPs and HOFIs were held for at least 2 weeks for acclimation to cage conditions prior to inoculation with recombinant viruses. AMCRs were captured by cannon net in Bellvue, CO, USA, whilst HOSPs and HOFIs were collected using grain-baited ground traps and mist nets in Bakersfield, CA, USA. Samples of 0.2 and 0.1 ml blood were drawn by jugular venipuncture from AMCRs and HOSPs/HOFIs, respectively, and serum collected in serum separator tubes was tested for the presence of neutralizing antibodies to WNV and SLEV by a 90% plaque reduction neutralization test, as described previously (Brault et al., 2004). Groups of eight AMCRs and six HOSPs or HOFIs were inoculated subcutaneously in the breast region with 1500 p.f.u. of each virus using a 27-gauge needle. Blood was collected from birds daily by jugular venipuncture up to 7 days p.i. Whole blood was added to diluent (DMEM with 20 % FBS) at a dilution of 1 : 10, allowed to coagulate at room temperature for 30 min and centrifuged at 2500 g for 10 min. The sera were stored at −80 °C until titrated for infectious units in Vero cells by plaque assay.
Avian viral load quantification and plaque size evaluation.
Daily blood samples were assayed by plaque formation on monolayers of Vero cells. In addition, avian inocula for all viruses were back titrated by plaque assay to confirm the uniformity of the doses administered. Briefly, serial tenfold dilutions of serum were inoculated onto Vero cells that were overlaid as described previously (Brault et al., 2004). Plaques were enumerated at 72 h p.i. and multiplied by the dilution factor to determine viral titres (ml serum)−1. An initial 1 : 10 dilution of serum, as well as the use of 200 µl of the lowest dilution, resulted in a limit of detection of 1.7 log10 p.f.u. ml−1. Plaque size determination was performed at 72 h p.i. by measuring the diameter of ten well-delineated viral plaques.
Sequencing.
All full-length consensus sequences were generated using primer sets described previously (Lanciotti et al., 1999). Briefly, total viral RNA from 140 µl culture supernatant was extracted using a QIAamp Viral RNA Mini kit according to the manufacturer’s instructions (Qiagen) and eluted in 80 µl nuclease-free water. Amplicons (~2.5 kb each) were generated using a one-step RT-PCR kit (Invitrogen), purified by gel extraction (Qiagen) and used for direct sequencing on an ABI 3130x Genetic Analyser. WNV RNA was extracted from AMCR serum samples (4 or 5 days p.i.) and brain tissue at time of death/euthanasia. RT-PCR was used to generate amplicons using primers that flanked the prM-141 and E-156 loci (Ke301: 5′-TGGAGAGGTGTGAACAAACAAAC-3′, WNc2206: 5′-CTTTGAGGGTGGTTGTAAAGG-3′) and sequenced to confirm the genetic identity at these positions following growth in AMCRs. All prM–E amplicons (~2 kb) generated from AMCR samples were compared with the original bird inoculum consensus sequence to assess genetic identity.
Statistical analyses.
Statistical analyses were performed on the duration of viraemia and peak viraemia for all experimental bird samples by analyses of variance. Multiple comparisons (i.e. confidence intervals for the difference of means) were performed using Tukey’s HSD adjustment for comparisons of means. A level of significance of P<0.05 was used for all comparisons.
Supplementary Material
Acknowledgements
Vincent Martinez and Brian Carroll provided invaluable avian collection and animal husbandry support for this project. Funding for these studies was provided by grants from the National Institutes of Health (AI061822 and AI55606) and the Centers for Disease Control and Prevention grant (CI000235). Trapping of AMCRs and HOFIs was performed under US Fish and Wildlife Scientific Collecting Permit MB-032526 and MB-082812 and Resident Scientific Collection Permit 801049-02 by the State of California Department of Fish and Game. Collection, husbandry and experimental inoculations of birds were performed under approved UC Davis IACUC protocols.
Footnotes
A supplementary figure is available with the online version of this paper.
References
- Adrián Diaz L., Komar N., Visintin A., Dantur Juri M. J., Stein M., Lobo Allende R., Spinsanti L., Konigheim B., Aguilar J., et al. & other authors (2008). West Nile virus in birds, Argentina. Emerg Infect Dis 14, 689–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrera R., Hunsperger E., Muñoz-Jordán J. L., Amador M., Diaz A., Smith J., Bessoff K., Beltran M., Vergne E., et al. & other authors (2008). First isolation of West Nile virus in the Caribbean. Am J Trop Med Hyg 78, 666–668 [PubMed] [Google Scholar]
- Beasley D. W., Davis C. T., Estrada-Franco J., Navarro-Lopez R., Campomanes-Cortes A., Tesh R. B., Weaver S. C., Barrett A. D. (2004). Genome sequence and attenuating mutations in West Nile virus isolate from Mexico. Emerg Infect Dis 10, 2221–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beasley D. W., Whiteman M. C., Zhang S., Huang C. Y., Schneider B. S., Smith D. R., Gromowski G. D., Higgs S., Kinney R. M., Barrett A. D. (2005). Envelope protein glycosylation status influences mouse neuroinvasion phenotype of genetic lineage 1 West Nile virus strains. J Virol 79, 8339–8347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch I., Herrera F., Navarro J. C., Lentino M., Dupuis A., Maffei J., Jones M., Fernández E., Pérez N., et al. & other authors (2007). West Nile virus, Venezuela. Emerg Infect Dis 13, 651–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brault A. C., Langevin S. A., Bowen R. A., Panella N. A., Biggerstaff B. J., Miller B. R., Komar N. (2004). Differential virulence of West Nile strains for American crows. Emerg Infect Dis 10, 2161–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brault A. C., Langevin S. A., Fang Y., Beasley D. W. C., Barker C. M., Sanders T., Reisen W. K., Barrett A. D. T., Bowen R. A. (2011). Reduced avian virulence and viremia response of West Nile viral isolates from Mexico and Texas. Am J Trop Med Hyg 85, 758–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis C. T., Beasley D. W., Guzman H., Siirin M., Parsons R. E., Tesh R. B., Barrett A. D. (2004). Emergence of attenuated West Nile virus variants in Texas, 2003. Virology 330, 342–350 [DOI] [PubMed] [Google Scholar]
- Davis C. T., Ebel G. D., Lanciotti R. S., Brault A. C., Guzman H., Siirin M., Lambert A., Parsons R. E., Beasley D. W., et al. & other authors (2005). Phylogenetic analysis of North American West Nile virus isolates, 2001–2004: evidence for the emergence of a dominant genotype. Virology 342, 252–265 [DOI] [PubMed] [Google Scholar]
- Deardorff E., Estrada-Franco J., Brault A. C., Navarro-Lopez R., Campomanes-Cortes A., Paz-Ramirez P., Solis-Hernandez M., Ramey W. N., Davis C. T., et al. & other authors (2006). Introductions of West Nile virus strains to Mexico. Emerg Infect Dis 12, 314–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohm D. J., Turell M. J. (2001). Effect of incubation at overwintering temperatures on the replication of West Nile Virus in New York Culex pipiens (Diptera: Culicidae). J Med Entomol 38, 462–464 [DOI] [PubMed] [Google Scholar]
- Dohm D. J., O’Guinn M. L., Turell M. J. (2002). Effect of environmental temperature on the ability of Culex pipiens (Diptera: Culicidae) to transmit West Nile virus. J Med Entomol 39, 221–225 [DOI] [PubMed] [Google Scholar]
- Dupuis A. P., II, Marra P. P., Kramer L. D. (2003). Serologic evidence of West Nile virus transmission, Jamaica, West Indies. Emerg Infect Dis 9, 860–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis A. P., II, Marra P. P., Reitsma R., Jones M. J., Louie K. L., Kramer L. D. (2005). Serologic evidence for West Nile virus transmission in Puerto Rico and Cuba. Am J Trop Med Hyg 73, 474–476 [PubMed] [Google Scholar]
- Eidson M. (2001). “Neon needles” in a haystack: the advantages of passive surveillance for West Nile virus. Ann N Y Acad Sci 951, 38–53 [PubMed] [Google Scholar]
- Elizondo-Quiroga D., Davis C. T., Fernandez-Salas I., Escobar-Lopez R., Velasco Olmos D., Soto Gastalum L. C., Aviles Acosta M., Elizondo-Quiroga A., Gonzalez-Rojas J. I., et al. & other authors (2005). West Nile Virus isolation in human and mosquitoes, Mexico. Emerg Infect Dis 11, 1449–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada-Franco J. G., Navarro-Lopez R., Beasley D. W., Coffey L., Carrara A. S., Travassos da Rosa A., Clements T., Wang E., Ludwig G. V., et al. & other authors (2003). West Nile virus in Mexico: evidence of widespread circulation since July 2002. Emerg Infect Dis 9, 1604–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezenwa V. O., Godsey M. S., King R. J., Guptill S. C. (2006). Avian diversity and West Nile virus: testing associations between biodiversity and infectious disease risk. Proc Biol Sci 273, 109–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y., Reisen W. K. (2006). Previous infection with West Nile or St. Louis encephalitis viruses provides cross protection during reinfection in house finches. Am J Trop Med Hyg 75, 480–485 [PubMed] [Google Scholar]
- Farfán-Ale J. A., Blitvich B. J., Loroño-Pino M. A., Marlenee N. L., Rosado-Paredes E. P., García-Rejón J. E., Flores-Flores L. F., Chulim-Perera L., López-Uribe M., et al. & other authors (2004). Longitudinal studies of West Nile virus infection in avians, Yucatán State, México. Vector Borne Zoonotic Dis 4, 3–14 [DOI] [PubMed] [Google Scholar]
- Farfán-Ale J. A., Blitvich B. J., Marlenee N. L., Loroño-Pino M. A., Puerto-Manzano F., García-Rejón J. E., Rosado-Paredes E. P., Flores-Flores L. F., Ortega-Salazar A., et al. & other authors (2006). Antibodies to West Nile virus in asymptomatic mammals, birds, and reptiles in the Yucatan Peninsula of Mexico. Am J Trop Med Hyg 74, 908–914 [PubMed] [Google Scholar]
- Farfán-Ale J. A., Loroño-Pino M. A., Garcia-Rejon J. E., Hovav E., Powers A. M., Lin M., Dorman K. S., Platt K. B., Bartholomay L. C., et al. & other authors (2009). Detection of RNA from a novel West Nile-like virus and high prevalence of an insect-specific flavivirus in mosquitoes in the Yucatan Peninsula of Mexico. Am J Trop Med Hyg 80, 85–95 [PMC free article] [PubMed] [Google Scholar]
- Garcia-Tapia D., Hassett D. E., Mitchell W. J., Jr, Johnson G. C., Kleiboeker S. B. (2007). West Nile virus encephalitis: sequential histopathological and immunological events in a murine model of infection. J Neurovirol 13, 130–138 [DOI] [PubMed] [Google Scholar]
- Gibbs S. E., Allison A. B., Yabsley M. J., Mead D. G., Wilcox B. R., Stallknecht D. E. (2006). West Nile virus antibodies in avian species of Georgia, USA: 2000–2004. Vector Borne Zoonotic Dis 6, 57–72 [DOI] [PubMed] [Google Scholar]
- Gubler D. J. (2007). The continuing spread of West Nile virus in the western hemisphere. Clin Infect Dis 45, 1039–1046 [DOI] [PubMed] [Google Scholar]
- Guptill S. C., Julian K. G., Campbell G. L., Price S. D., Marfin A. A. (2003). Early-season avian deaths from West Nile virus as warnings of human infection. Emerg Infect Dis 9, 483–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz F. X., Allison S. L. (2000). Structures and mechanisms in flavivirus fusion. Adv Virus Res 55, 231–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz F. X., Auer G., Stiasny K., Holzmann H., Mandl C., Guirakhoo F., Kunz C. (1994). The interactions of the flavivirus envelope proteins: implications for virus entry and release. Arch Virol Suppl 9, 339–348 [DOI] [PubMed] [Google Scholar]
- Johnson P. T., Thieltges D. W. (2010). Diversity, decoys and the dilution effect: how ecological communities affect disease risk. J Exp Biol 213, 961–970 [DOI] [PubMed] [Google Scholar]
- Johnson G., Nemeth N., Hale K., Lindsey N., Panella N., Komar N. (2010). Surveillance for West Nile virus in American white pelicans, Montana, USA, 2006–2007. Emerg Infect Dis 16, 406–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney R. M., Huang C. Y., Whiteman M. C., Bowen R. A., Langevin S. A., Miller B. R., Brault A. C. (2006). Avian virulence and thermostable replication of the North American strain of West Nile virus. J Gen Virol 87, 3611–3622 [DOI] [PubMed] [Google Scholar]
- Komar N. (2001). West Nile virus surveillance using sentinel birds. Ann N Y Acad Sci 951, 58–73 [DOI] [PubMed] [Google Scholar]
- Komar N., Clark G. G. (2006). West Nile virus activity in Latin America and the Caribbean. Rev Panam Salud Publica 19, 112–117 [DOI] [PubMed] [Google Scholar]
- Komar O., Robbins M. B., Contreras G. G., Benz B. W., Klenk K., Blitvich B. J., Marlenee N. L., Burkhalter K. L., Beckett S., et al. & other authors (2005). West Nile virus survey of birds and mosquitoes in the Dominican Republic. Vector Borne Zoonotic Dis 5, 120–126 [DOI] [PubMed] [Google Scholar]
- Lanciotti R. S., Roehrig J. T., Deubel V., Smith J., Parker M., Steele K., Crise B., Volpe K. E., Crabtree M. B., et al. & other authors (1999). Origin of the West Nile virus responsible for an outbreak of encephalitis in the northeastern United States. Science 286, 2333–2337 [DOI] [PubMed] [Google Scholar]
- Li L., Lok S.-M., Yu I.-M., Zhang Y., Kuhn R. J., Chen J., Rossmann M. G. (2008). The flavivirus precursor membrane–envelope protein complex: structure and maturation. Science 319, 1830–1834 [DOI] [PubMed] [Google Scholar]
- Mattar S., Edwards E., Laguado J., González M., Alvarez J., Komar N. (2005). West Nile virus antibodies in Colombian horses. Emerg Infect Dis 11, 1497–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra M., Schein C. H. (2007). Flavitrack: an annotated database of flavivirus sequences. Bioinformatics 23, 2645–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Betoulle M. E., Morales H., Blitvich B. J., Powers A. M., Davis E. A., Klein R., Cordón-Rosales C. (2006). West Nile virus in horses, Guatemala. Emerg Infect Dis 12, 1038–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostashari F., Kulldorff M., Hartman J. J., Miller J. R., Kulasekera V. (2003). Dead bird clusters as an early warning system for West Nile virus activity. Emerg Infect Dis 9, 641–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson S., Jost C. A., Xu Q., Ess J., Martin J. E., Oliphant T., Whitehead S. S., Durbin A. P., Graham B. S., et al. & other authors (2008). Maturation of West Nile virus modulates sensitivity to antibody-mediated neutralization. PLoS Pathog 4, e1000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesko K., Mores C. N. (2009). Effect of sequential exposure on infection and dissemination rates for West Nile and St. Louis encephalitis viruses in Culex quinquefasciatus. Vector Borne Zoonotic Dis 9, 281–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L. M., Johansson M. A., Panella N., McLean R., Creekmore T., Puelle R., Komar N. (2009). Declining mortality in American crow (Corvus brachyrhynchos) following natural West Nile virus infection. Avian Dis 53, 458–461 [DOI] [PubMed] [Google Scholar]
- Reisen W. K., Fang Y., Martinez V. M. (2005). Avian host and mosquito (Diptera: Culicidae) vector competence determine the efficiency of West Nile and St. Louis encephalitis virus transmission. J Med Entomol 42, 367–375 [DOI] [PubMed] [Google Scholar]
- Reisen W. K., Lothrop H. D., Wheeler S. S., Kennsington M., Gutierrez A., Fang Y., Garcia S., Lothrop B. (2008). Persistent West Nile virus transmission and the apparent displacement St. Louis encephalitis virus in southeastern California, 2003–2006. J Med Entomol 45, 494–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Miyoshi H., Goto A., Ako Y., Ueki T., Kariwa H., Takashima I. (2004). Viral envelope protein glycosylation is a molecular determinant of the neuroinvasiveness of the New York strain of West Nile virus. J Gen Virol 85, 3637–3645 [DOI] [PubMed] [Google Scholar]
- Silva M. C., Guerrero-Plata A., Gilfoy F. D., Garofalo R. P., Mason P. W. (2007). Differential activation of human monocyte-derived and plasmacytoid dendritic cells by West Nile virus generated in different host cells. J Virol 81, 13640–13648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele K. E., Linn M. J., Schoepp R. J., Komar N., Geisbert T. W., Manduca R. M., Calle P. P., Raphael B. L., Clippinger T. L., et al. & other authors (2000). Pathology of fatal West Nile virus infections in native and exotic birds during the 1999 outbreak in New York City, New York. Vet Pathol 37, 208–224 [DOI] [PubMed] [Google Scholar]
- Stiasny K., Heinz F. X. (2006). Flavivirus membrane fusion. J Gen Virol 87, 2755–2766 [DOI] [PubMed] [Google Scholar]
- Swaddle J. P., Calos S. E. (2008). Increased avian diversity is associated with lower incidence of human West Nile infection: observation of the dilution effect. PLoS ONE 3, e2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wünschmann A., Shivers J., Carroll L., Bender J. (2004). Pathological and immunohistochemical findings in American crows (Corvus brachyrhynchos) naturally infected with West Nile virus. J Vet Diagn Invest 16, 329–333 [DOI] [PubMed] [Google Scholar]
- Zhang W., Chipman P. R., Corver J., Johnson P. R., Zhang Y., Mukhopadhyay S., Baker T. S., Strauss J. H., Rossmann M. G., Kuhn R. J. (2003). Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat Struct Biol 10, 907–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.