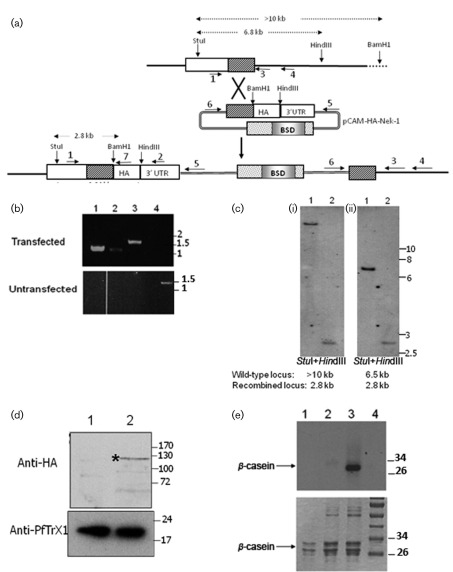
Fig. 3.
HA tagging of the pfnek-1 locus. (a) Strategy for HA tagging. The locations of the primers used in PCR diagnostics are indicated with numbered arrows, and the positions of the restriction sites used for Southern blot analysis are shown. (b) PCR genotyping analysis. Genomic DNA was isolated from pCAM-HA-nek-1-transfected parasites and from parental 3D7 parasites, and subjected to PCR using the indicated primers (see Fig. 3a). Lanes: 1, primers 1+7 (diagnostic for 5′ integration event, 1.2 kb); 2, primers 6+3 (diagnostic for 3′ integration event, 1.2 kb); 3, primers 6+4 (diagnostic for 3′ integration event, 1.5 kb); 4, primers 1+4 (diagnostic for wild-type locus, 1.5 kb). Sizes of co-migrating markers are indicated in kb. (c) Southern blot analysis of transfected parasites. Total DNA digested with StuI and HindIII (i) or StuI and BamHI (ii) was probed with a fragment spanning nucleotides 577–1155 of the pfnek-1 coding region. Sizes of co-migrating markers are indicated in kb. (d) Western blot analysis of HA-tagged Pfnek-1. Protein extracts (15 µg) from the parental 3D7 wild-type clone (lane 1) and pCAM-HA-nek-1-transfected parasites (lane 2) were probed with an anti-HA antibody. Sizes of co-migrating markers are indicated in kDa. (e) Anti-HA antibodies immunoprecipitate kinase activity from transfected parasites. Anti-HA antibodies were incubated with extracts from the parental 3D7 wild-type clone (lane 2) or pCAM-HA-Nek-1-transfected parasites (lane 3). Immunoprecipitated material was assayed for β-casein activity. Lane 1 shows the kinase assay with β-casein alone (no immunoprecipitate added), and the molecular mass marker is shown in lane 4. Molecular masses are indicated in kDa.