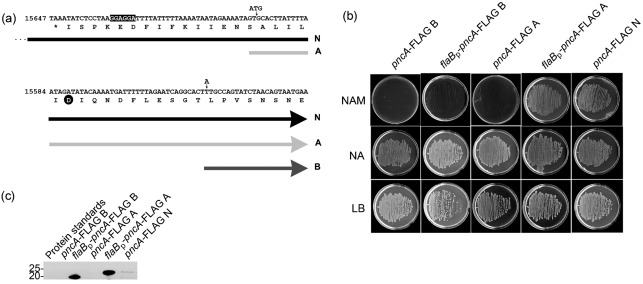
Fig. 1.
Identification of the B. burgdorferi pncA sequence sufficient to encode a functional nicotinamidase enzyme. (a) Schematic representation of the three pncA-FLAG constructs used for functional complementation of an S. typhimurium pncAnadB mutant for growth on minimal medium containing 0.1 mM nicotinamide. The reverse complement of nt 15678–15558 on lp25 is shown, with the translated single-letter amino acids indicated below. The 5′ sequences of each pncA construct are represented by block arrows: pncA-FLAG N (N, black arrow; note that the 5′ end of this construct continues approximately 900 bp upstream of the sequence that is shown, as indicated by three dots), pncA-FLAG A (A, light-grey arrow) and pncA-FLAG B (B, dark-grey arrow). The putative ribosome-binding site sequence is highlighted in black. The aspartic acid residue conserved among the catalytic triad of PncA homologues is indicated by a black circle. The locations of the engineered ATG start codon for construct pncA-FLAG A, and the nucleotide point mutation (T to A) to create an ATG start codon for construct pncA-FLAG B, are indicated by small vertical arrows. (b) Functional-complementation assay for nicotinamidase activity of pncA-FLAG constructs. Representative data are shown for the overnight growth at 37 °C of the S. typhimurium pncAnadB mutant harbouring each pncA-FLAG construct (indicated above each column) on minimal agar containing 0.1 mM nicotinamide (NAM) or 0.1 mM nicotinic acid (NA). Rich agar (LB) was used as a positive control. All agar plates contained 50 µg kanamycin ml−1. (c) Immunoblot analysis of PncA–FLAG production in the S. typhimurium pncAnadB mutant harbouring each pncA-FLAG gene. Total protein lysates were resolved by SDS-PAGE, transferred to a nitrocellulose membrane and probed for PncA–FLAG using an anti-FLAG M2 monoclonal antibody. Sizes of the protein standards are indicated in kDa. The predicted molecular masses for PncA–FLAG B and PncA–FLAG A are 21 and 24 kDa, respectively.