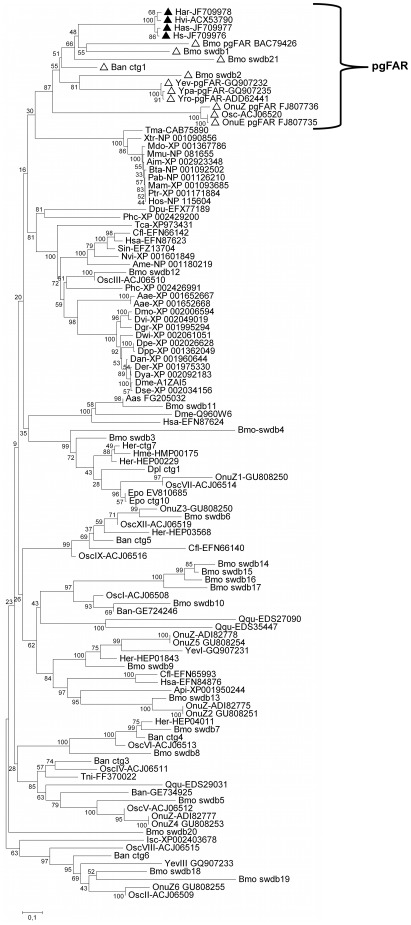
Figure 3. FAR gene tree.
Gene tree of arthropod, mammalian and lepidopteran FARs including the lepidopteran–specific pgFAR group, supported with a bootstrap value of 87 and marked with a bracket. Within this group, heliothine pgFARs are marked with dark triangles and other biosynthetic moth reductases with transparent triangles. The Neighbor–joining algorithm analysis was computed in MEGA (v. 4.0) using deduced aa sequences with pairwise deletion, and the JTT matrix based model with 1,500 bootstrap replicates. Sequences were retrieved from GenBank by manually searching for arthropod FARs, as well as BLASTP database searches using HvFAR as query. B. mori FAR sequences were retrieved from the Silkworm Genome Database. All sequences noted as cng (contig) were obtained from Liénard et al. [15]. The full species names can be found in (Table S2). Sequences were aligned with the ClustalW2 algorithm with the ClustalX2 interface and manually inspected before computing the phylogenetic relationship.