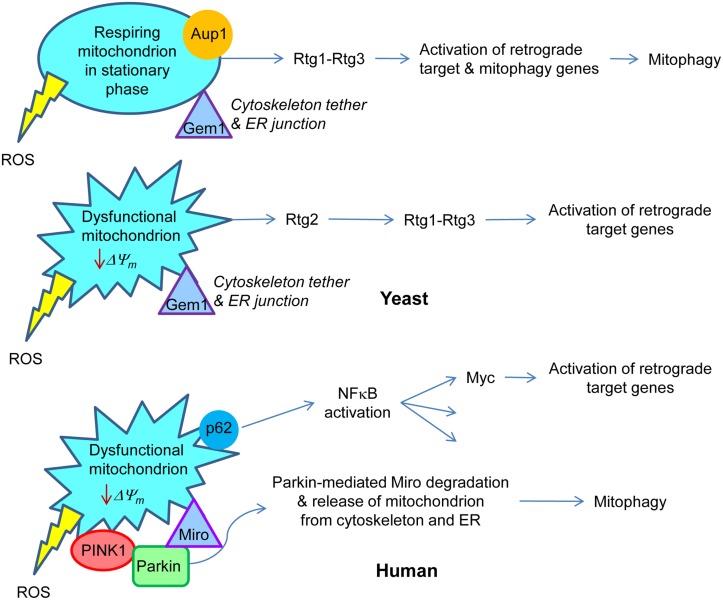
Figure 2.
Retrograde signaling in yeast and human. In yeast, respiring mitochondria in non-dividing, stationary phase cells signal the retrograde response that activates both retrograde response target genes, similar to those in dividing cells, and mitophagy genes. This results in the metabolic adaptation to stationary phase. Aup1, a protein phosphatase in the intermembrane space in mitochondria, is essential for this gene induction. Rtg1–Rtg3 is the retrograde transcription factor. On the other hand, dysfunctional mitochondria in growing cells trigger the classical retrograde response with activation of retrograde response target genes. Rtg2 plays an essential role in this process. Gem1 is a Miro homolog in yeast which is important for maintaining junctions between mitochondria and the endoplasmic reticulum. By analogy with mammalian cells, it would also tether the mitochondria to the cytoskeleton. In human cells, a drop in mitochondrial membrane potential (ΔΨm) recruits Parkin by the PINK1 protein kinase to the mitochondrial membrane. Parkin mediates ubiquitylation of Miro, which releases the mitochondria from the cytoskeleton and also, presumably, from the endoplasmic reticulum. This facilitates the removal of dysfunctional mitochondria by mitophagy. Sequestosome 1 (p62) aggregates proteins polyubiquitinated by Parkin on the surface of mitochondria. p62 is known to stimulate NFκB, which among its many target genes has Myc. The Myc–Max dimer is homologous to Rtg1–Rtg3. Transcription of Myc is activated in human cells devoid of mtDNA, and Myc itself activates the transcription of metabolic genes, typical for the retrograde response. The production of reactive oxygen species (ROS) by the mitochondria may elicit responses as well.