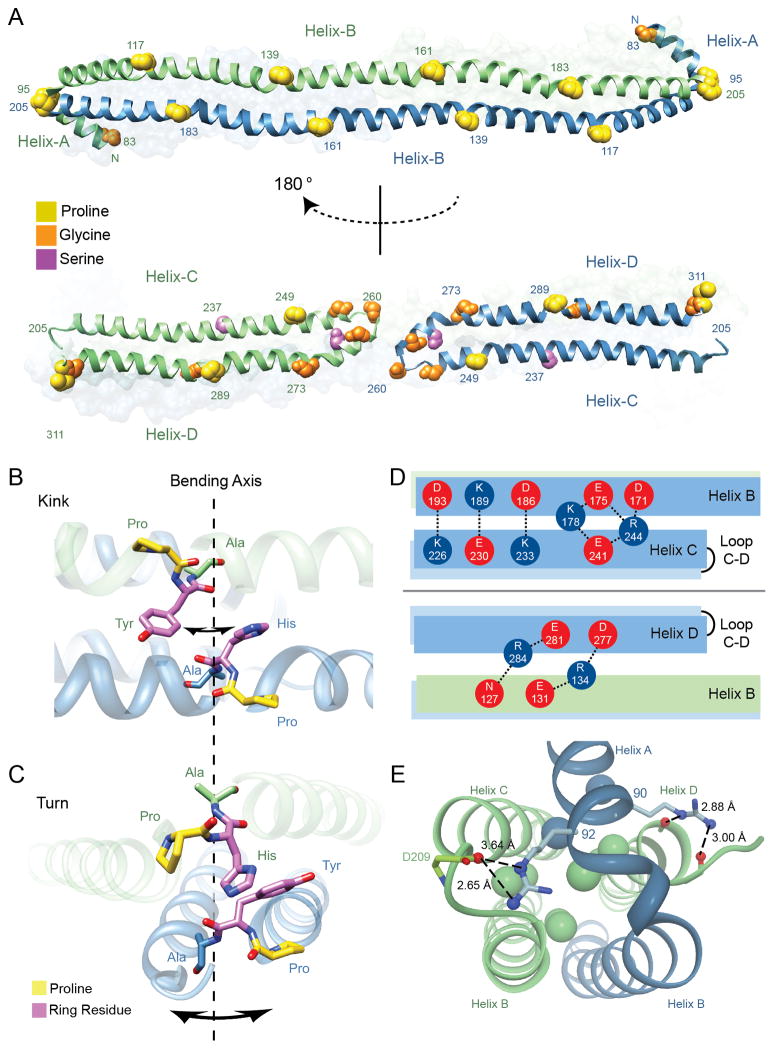
Figure 3.
Molecular characteristics of the apoA-IV64-335 dimer. (A) Alignment of helix disrupting ‘kinks’ reveals paired segmentation. Proline, glycine and serine residues are represented by spheres and colored accordingly. (top) Helix-B from both green and blue chains run anti-parallel and conserved proline residues from each helix are in a shifted alignment, dividing the B helices into five paired segments of 22 a.a. in length. (bottom) Segmentation is also observed between Helix-C and D, where proline residues align vertically with glycine and serine. Additional glycine residues that form the Helix-C to D turn are also displayed with the central glycine (Gly 260) annotated. (B) Illustration of the P(H/Y)A motifs that align vertically across Helices-B looking down onto the two stacked helices (hydrophobic core at the top and polar exterior on the bottom). Shown are residues PHA (161-163) and PYA (139-141) from opposing B helices. The potential axis of bending and direction is indicated with arrows around dashed line. (C) Illustration of paired P(H/Y)A motifs that align in turns (95-97 linking Helix-A to B) and (205-207 linking opposing Helix-B to C). Orientation is looking down the length of the dimer rod. (D) Hydrogen bonding and salt bridges at the interface of Helix-B and helical arms (Helix-C and D). (top) Intramolecular interactions are shown between acidic (red) and basic (blue) residues looking down on the dimer rod parallel to the plane made up by the stacked B-helices. (bottom) Intermolecular interactions are shown looking up in the same plane (i.e. rotated 180°). (E) Helix-A caps the four-helix bundle at each opposing dimer end. Shown are interactions of two conserved arginine residues (90 and 92) within Helix-A that interact at the top of the bundle. Interior leucine residues are shown as spheres and colored in accordance with each monomer. See also Figures S3, S4 and S8, and Table S2.