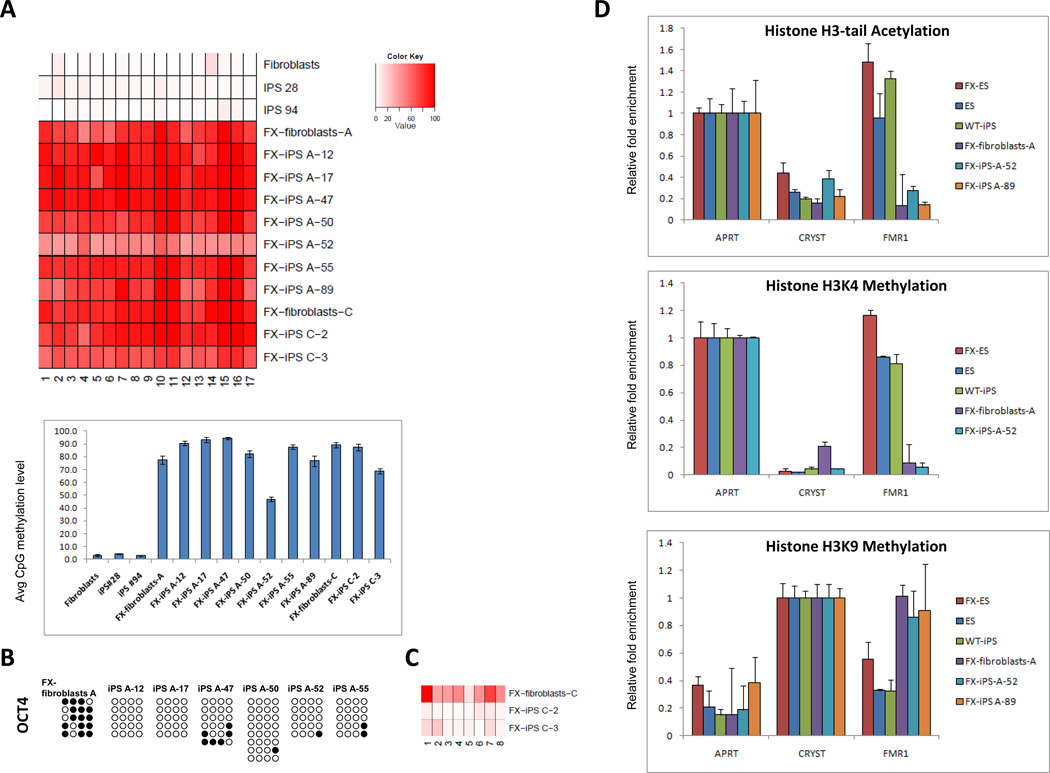
Figure 2. Epigenetic modifications in FX-iPS cells.
A. Pyrosequencing analysis of the FMR1 promoter in wt-fibroblasts, two clones of wt-iPS cells (iPS 28, iPS 94), FX-fibroblasts-A, and seven derivative iPS cell lines (FX-iPS A-12, 17, 47, 50, 52, 55, 89), FX-fibroblasts-C and two derivative iPS cell lines (FX-iPS C-2, C-3). Upper panel – methylation level at CpG position. Bottom panel – average methylation level of all CpG sites for each sample. B. Bisulfite sequencing analysis of the OCT4 promoter in FX-fibroblasts-A and FX-iPS cell clones derived from them. Open circles, unmethylated CpGs; Black circles, methylated CpGs. C. Pyrosequencing analysis of the OCT4 5’UTR in FX-fibroblasts-C and FX-iPS cell clones derived from them. D. Histone modifications at the FMR1 locus in FX-iPS cells: ChIP analysis of histone H3-tail acetylation and H3K4 and H3K9 methylation in FX-iPS cells. Real-time PCR was performed on bound and input sonicated DNA fragments using primers for the FMR1 promoter. Adenine phosphoribosyl transferase (APRT) and Crystalline (CRYST) served as positive and negative controls, respectively. Values were normalized to the appropriate positive control and shown with their respective standard errors.