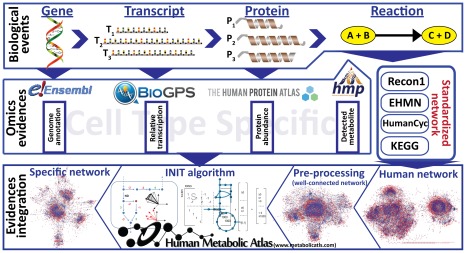
Figure 1. General pipeline used in the reconstruction of cell specific genome-scale metabolic networks.
Biological information at the genome, transcriptome, proteome and metabolome levels contained in publicly available databases and generic human GEMs (Recon1, EHMN, HumanCyc) is integrated to form a generic human metabolic network, which is processed in order to obtain the connected iHuman1512 network. Subsequently, the cell type specific evidence is used to generate cell type specific subnetworks using the INIT algorithm.