Abstract
We analyzed 40 SNP and 19 STR Y-chromosomal markers in a large sample of 1,525 indigenous individuals from 14 populations in the Caucasus and 254 additional individuals representing potential source populations. We also employed a lexicostatistical approach to reconstruct the history of the languages of the North Caucasian family spoken by the Caucasus populations. We found a different major haplogroup to be prevalent in each of four sets of populations that occupy distinct geographic regions and belong to different linguistic branches. The haplogroup frequencies correlated with geography and, even more strongly, with language. Within haplogroups, a number of haplotype clusters were shown to be specific to individual populations and languages. The data suggested a direct origin of Caucasus male lineages from the Near East, followed by high levels of isolation, differentiation and genetic drift in situ. Comparison of genetic and linguistic reconstructions covering the last few millennia showed striking correspondences between the topology and dates of the respective gene and language trees, and with documented historical events. Overall, in the Caucasus region, unmatched levels of gene-language co-evolution occurred within geographically isolated populations, probably due to its mountainous terrain.
Keywords: Y chromosome, glottochronology, Caucasus, gene geography
INTRODUCTION
Since the Upper Paleolithic, anatomically modern humans have been present in the Caucasus region, which is located between the Black and Caspian Seas at the boundary between Europe and Asia. While Neolithization in the Transcaucasus (south Caucasus) was stimulated by direct example and possible in-migration from the Near East, in the North Caucasus archaeologists have stressed the cultural succession from the Upper Paleolithic to the Mesolithic and Neolithic (Bader and Tseretely, 1989; Bzhania, 1996). Neolithic cultures developed in the North Caucasus ~7,500 years before present (YBP) from the local Mesolithic cultures (microlithic stone industries indicate a gradual transition), and, once established, domesticated local barley and wheat species (Masson et al., 1994; Bzhania, 1996). Only in the Early Bronze Age (5,200-4,300 YBP) did cultural innovations from the Near East become more intensive with the emergence of the Maikop archaeological culture (Munchaev, 1994). These could have occurred alongside migratory events from the area between the Tigris River in the east and northern Syria and adjacent East Anatolia in the west (Munchaev, 1994: 170). The Late Bronze Age Koban culture (3,200-2,400 YBP), which predominated across the North Caucasus until Sarmatian times (2,400-2,300 YBP), became the common cultural substrate for most of the present-day peoples in the North Caucasus (Melyukova, 1989: 295).
After approximately 1,500 YBP, this pattern changed, and most migration came to the North Caucasus from the East European steppes to the north, rather than from the Near East to the south. These new migrants were Iranian speakers (Scythians, Sarmatians and their descendants, the Alans) who arrived around 3,000-1,500 YBP, followed by Turkic speakers about 500-1,000 YBP. The new migrants forced the indigenous Caucasian population to relocate from the foothills into the high mountains. Some of the incoming steppe dwellers also migrated to the highlands, mixing with the indigenous groups and acquiring a sedentary lifestyle (Abramova, 1989; Melyukova, 1989; Ageeva, 2000). The defeat of the Alans by the Mongols, and then by Tamerlane in the 14th century, stimulated the expansion of the indigenous populations of the West Caucasus into former Alan lands (Fedorov, 1983). A later expansion of Turkic-speaking Nogais took place about 400 YBP.
The genetic contributions of these three components (indigenous Upper Paleolithic settlement; Bronze Age Near Eastern expansions; Iron Age migration from the steppes) is unknown. Physical anthropologists have traced the continuity of local anthropological (cranial) types from the Upper Paleolithic, but data reflecting the influences from the Near East and Eastern Europe are contradictory (Abdushelishvili, 1964; Alexeev, 1974; Gerasimova, Rud’, and Yablonskij, 1987).
Linguistically, the North Caucasus is a mosaic consisting of more than 50 languages, most of which belong to the North Caucasian language family. Iranian languages (Indo-European family) are represented by Ossets, and there are a variety of Turkic-speaking groups (Altaic family) as well (Ruhlen, 1987). A number of studies of North Caucasian languages (Trubetzkoy, 1930; Gigeneishvili, 1977; Shagirov, 1977; Talibov, 1980; Bokarev, 1981; Chirikba, 1996) collected linguistic data and established regular phonetic correspondences between North Caucasian languages. The resulting classification (Nikolaev and Starostin, 1994) became generally accepted with some modifications (Kuipers, 1963; Comrie, 1987; Ruhlen, 1987; www.ethnologue.com).
This classification was based on the common innovation method and particularly on the glotto-chronological method (Starostin, 1989), which is now widely used by the Evolution of Human Languages Project coordinated by the Santa Fe Institute (http://ehl.santafe.edu/intro1.htm, http://starling.rinet.ru/main.html). For example, application of this method to the modern Romance languages (Spanish, Italian, Romanian, etc.) produced a date for their split to about 1,600 years BP. This date corresponds to the time of the disintegration of the Roman Empire, when Latina Vulgata (the common language in the Empire’s provinces) became subdivided into regional dialects (Blazhek and Novotna, 2008). Thus, this result validates the methodology, at least for this example.
The present work employs Starostin’s methodology, and we made special efforts to create the high-quality linguistic databases required for this analysis. Thus, based on significantly extended and revised linguistic databases, we have applied a glotto-chronological approach to the North Caucasian languages. As a result, our study provides a unique opportunity to make direct comparisons of linguistic and genetic data from the same populations. Lexico-statistical methods have also been applied to a number of language families using a Bayesian approach to increase the statistical robustness of language classification (Gray and Atkinson, 2003; Kitchen et al., 2009; Greenhill et al., 2010). Using these methods with the Caucasus languages under study here will be the focus of future work.
Previous studies of genetic diversity in the Caucasus (Nasidze et al., 2003, 2004a, b) noted that geography, rather than language, provides a better (but statistically non-significant) explanation for the observed genetic structure. However, while the sample sizes for Y-chromosomal markers in those studies were large for southern populations, they were substantially smaller for North Caucasus populations (average n = 28, with the exception of the Ossets). Several subsequent papers have explored the genetic composition of the Dagestan in the east Caucasus (Bulaeva et al., 2006; Tofanelli et al., 2009; Caciagli et al., 2009). A later survey of the Y-chromosomal composition of the Caucasus was published in Russian only, with phylogenetic resolution no deeper than the designation of haplogroups G-M201, J1-M267 and J2-M172 (Kutuev et al., 2010). Some data can also be retrieved from papers that focused other regions (Rosser et al., 2000; Semino et al., 2000; Wells et al., 2001; Zerjal et al., 2002; Di Giacomo et al., 2004; Semino et al., 2004; Cruciani et al., 2007; Battaglia et al., 2009). To the best of our knowledge, no other Y-chromosome data from the Caucasus have been published, and, except for Georgians and the Ossets, reliable data sets are very few.
Our study presents a much more extensive survey of Y-chromosomal variation in the Caucasus. All geographic subregions are covered and all large ethnic groups are represented by large sample sizes (navg = 109). We did not include Turkic-speaking populations, as their recent immigration from Eastern Europe and Central Asia could possibly blur the deeper genetic patterns. Instead, we subtyped previously analyzed samples from the Near East (El-Sibai et al., 2009; Haber et al., 2010) and new samples from Eastern Europe for comparative purposes. Our genotyping strategy included the deepest known level of phylogenetic resolution for the common Caucasus haplogroups, as well as 19 STRs to facilitate the dating of these genetic lineages.
Overall, the present study sets out to draw a precise and reliable portrait of the Y-chromosomal and linguistic variation in the Caucasus, and to use this information to generate a more comprehensive history of the peoples of this area.
MATERIALS AND METHODS
Samples
A total of 1,525 blood samples from fourteen Caucasus populations (Table 1) were collected in 1998-2009 under the supervision of Elena Balanovska using a standardized sampling strategy. All sampled individuals identified their four grandparents as members of the given ethnic group, and were unrelated at least up to the third degree of relation. Informed consent was obtained under the control of the Ethics Committee of the Research Centre for Medical Genetics, Russia.
Table 1.
Characteristics of the study populations
| Population | N | Language group |
Geograp hic position – |
Region (country, province, district) |
Latitu de |
Longit ude |
Sampling supervisor(s) |
|---|---|---|---|---|---|---|---|
| Shapsug | 100 | Abkhazo- Adyghian |
West Caucasus |
Adygei republic, Tuapsinskii, Lazarevskii district |
44.15 | 39.12 | Balanovska |
| Abkhaz | 58 | Abkhazo- Adyghian |
West Caucasus |
Abkhazian republic | 43.10 | 41.10 | Balanovska, Pocheshkhova |
| Circassians | 142 | Abkhazo- Adyghian |
West Caucasus |
Karachai-Cherkess republic Habezskii, Prikubanskii district |
43.80 | 41.75 | Balanovska, Pocheshkhova |
| Ossets-Iron | 230 | Iranian | Сentral Caucasus |
North Ossetian republic Alagirskii, Irafskii, Prigorodnyi district |
42.90 | 44.47 | Balanovska |
| Ossets-Digor | 127 | Iranian | Сentral Caucasus |
North Ossetian republic Digorskii district |
43.12 | 43.55 | Balanovska |
| Ingush | 143 | Nakh | East Caucasus |
Ingushetia republic Nazran’ Malgobekskii |
43.12 | 45.04 | Balanovska, Pocheshkhova |
| Chechen (Ingushetia) |
112 | Nakh | East Caucasus |
Ingushetia republic Malgobekskii district |
43.20 | 45.20 | Balanovska, Pocheshkhova |
| Chechen (Chechnya) |
118 | Nakh | East Caucasus |
Chechnya republic Achhoi-Martanovskii district |
43.20 | 45.98 | Balanovska, Pocheshkhova |
| Chechen (Dagestan) |
100 | Nakh | East Caucasus |
Dagestan republic Hasavyurtovskii, Kazbekovskii, Novolakskii district |
43.32 | 46.55 | Balanovska, Radzhabov |
| Avar | 115 | Dagestan | East Caucasus |
Dagestan republic Shamil’skii, Buinakskii, Uncukul’skii, Gunibskii district |
42.47 | 46.88 | Balanovska |
| Dargins | 101 | Dagestan | East Caucasus |
Dagestan republic Akushinskii and Dahadaevskii districts |
42.18 | 47.22 | Balanovska |
| Kubachi | 65 | Dagestan | East Caucasus |
Dagestan republic Dahadaevskii district, Kubachi |
42.08 | 47.58 | Balanovska, Radzhabov |
| Kaitak | 33 | Dagestan | East Caucasus |
Dagestan republic Kaitakskii district |
42.15 | 47.63 | Balanovska, Radzhabov |
| Lezghins | 81 | Dagestan | East Caucasus |
Dagestan republic Ahtynskii district |
41.55 | 47.70 | Balanovska |
We also included 254 samples from the Near East (El-Sibai et al., 2009; Haber et al., 2010), which were further subtyped here for SNPs within haplogroups J2-M172 and G2a-P15.
Molecular Genetic Analysis
DNAs were extracted from white cells using an organic extraction method (Powell and Gannon, 2002). DNA concentration was evaluated via quantitative real-time PCR using the Quantifiler DNA Quantification Kit (Applied Biosystems) and normalized to 1 ng/μl. Samples were SNP-genotyped using the Applied Biosystems 7900HT Fast Real-Time PCR System with a set of 40 custom TaqMan assays (Applied Biosystems). The samples were additionally amplified at 19 Y-chromosomal STR loci in two multiplexes and read on an Applied Biosystems 3130xl Genetic Analyzer. The first multiplex was the 17 STR loci Y-filerTM PCR Amplification Kit (Applied Biosystems). The remaining two STR loci, DYS388 and DYS426, along with six insertion-deletion polymorphisms (M17, M60, M91, M139, M175, M186) were genotyped in a separate custom multiplex also provided by Applied Biosystems. Quality control procedures included checking SNP genotypes for phylogenetic consistency, comparing with the haplogroup predicted from STR profiles (http://www.hprg.com/hapest5/index.html), and independent replication of 20 samples at the University of Arizona.
Statistical Analysis
Haplogroup frequency maps were created with the GeneGeo software using algorithms described previously (Balanovsky et al., 2008). Nei’s genetic distances between populations were calculated using the DJ software (Balanovsky et al., 2008) and visualized using multidimensional scaling plots and tree diagrams constructed with Statistica 6.0 (StatSoft. Inc., 2001). Geographic distances between populations were obtained from geographic coordinates in DJ software using spherical formulae.
To estimate correlation and partial correlation coefficients between matrices of genetic, geographic and linguistic distances, we conducted Mantel tests using Arlequin 3.11 (Schneider et al., 2000). The same software was used for the hierarchical analysis of molecular variation (AMOVA). Genetic boundaries were identified by Barrier 2.2 software (Manni and Guerard, 2004).
Network Analysis
The phylogenetic relationships between haplotypes within a haplogroup were estimated with the Reduced Median (RM) network algorithm in the program Network 4.1.1.2 (Bandelt et al., 1995), with the reduction threshold equal to 1. RM networks were visualized with Network Publisher (Fluxus Engineering, Clare, U.K.). No available algorithm automatically identifies haplotype clusters within the network; doing this by hand (the common practice) is inevitably arbitrary to some degree. Therefore, we applied the following rules to minimize variation between individuals when identifying the clusters: i) Because all the networks used had a clear center (the probable root), we considered as clusters only those groups of haplotypes that were linked to the root via an individual nodal haplotype (put differently, monophyletic branches in the network); ii) This cluster-specific shared node was considered to be the founder haplotype (selecting the founder is important for the age calculation using the ρ estimator); iii) To avoid using small sample sizes, we only considered clusters consisting of 10 or more samples; iv) Finally, we required that a cluster should be highly (above 80%) specific to a given population or group of closely-related populations.
Dating Genetic Lineages
To estimate the age of particular Y-chromosomal lineages in Caucasus populations, we applied four commonly used methods. First, the ρ (rho) estimator (Forster et al., 1996; Saillard et al., 2000) was used to date haplotype clusters. Second, BATWING (Wilson et al., 2003) was used to obtain independent dates for these haplotype clusters. Third, BATWING was also used to estimate the possible sequence and dates of population splits. Fourth, the standard deviation (SD) estimator (Sengupta et al., 2006) was used to estimate the age required to accumulate the observed diversity within populations for entire haplogroups (not for clusters within the haplogroup as in the first and second analyses).
The Time to the Most Recent Common Ancestor (TMRCA) of the clusters of STR haplotypes that appeared to have evolved within specific populations, and which were identified in the networks, was estimated with the ρ statistic according to Saillard et al. (2000). Since haplotype clusters are population-specific, the resulting age estimations serve as lower bounds for the time that a population may have been isolated following a split.
BATWING provides a mechanism to identify bounds, established by coalescent events, between which a unique event polymorphism (UEP) may have emerged. UEPs are typically identified by SNPs, but can be generalized to include “virtual UEPs” that mark clusters of phylogenetically related STR haplotypes identified by Network, following the methods of Cruciani et al. (2004, 2006). Populations within which the clusters are observed were identified, and BATWING computations included all samples representing those populations in order to estimate UEP dates. Markers DYS385a and DYS385b were excluded from calculations. We used prior distribution parameters obtained from genealogical mutation rates reported in Ge et al. (2009). BATWING was configured to start with an ancestral effective population size that began an exponential expansion at a date that BATWING estimated.
BATWING was also employed to model the sequence of the population splits and to estimate the split times among populations. These estimates did not employ the virtual UEPs identified using Network. As such, this procedure provides an independent check for the times of the population splits.
Standard deviations (SD) of microsatellite variances for four major haplogroups, G2a3b1-P303, G2a1a-P18, J2a4b*-M67(xM92) and J1*-M267(xP58), were calculated for each population with a sample size of at least five individuals from a given haplogroup. The confidence interval was estimated based on the standard error of the SD. This method is based on average squared difference (ASD) in STR variation. It does not estimate the population divergence time but instead the age (amount of time) required to produce the observed microsatellite variation within the haplogroup at each population, under the assumption of limited gene flow. All haplotypes of the haplogroup were used in the calculations and not just specific clusters. Calculations were carried out both including and excluding DYS385a and DYS385b because these markers were not typed in a locus-specific manner in our data. In the majority of the cases, the inclusion or the exclusion of these markers did not influence the estimates. The DYS385 and DYS388 loci were thus only excluded from the reported values when the microsatellite variances associated with them were 10-fold higher than the average variance.
When using ρ and SD estimators, we applied both an evolutionary mutation rate (6.9 × 10−4 per locus per generation; Zhivotovsky et al. 2004) and a genealogical rate (2.1 × 10−3; Gusmao et al., 2005; Sanchez-Diz et al., 2008; Ge et al., 2009) to convert the observed variation into a number of generations. The results obtained by using these two rates were compared with linguistic and historical evidence. The BATWING prior distribution parameters were based on only the genealogical rate because BATWING models mutations in each generation of a genealogy and the genealogical rate seems therefore to be most suitable for this analysis. In all methods, when converting the number of generations into calendar years we had to use a different generation time for each: 25 years with the evolutionary rate because this rate was initially estimated in years and converted into generations using 25 years/generation (Zhivotovsky et al., 2004), and 30 years for the genealogical rate because this is the approximate male generation time measured in demographic and anthropological studies (Fenner, 2005) and shown for the Caucasus populations in the genetico-demographic study (Pocheshkhova, 2008).
Dating Languages
Linguistic dates were calculated from the number of word substitutions that have accumulated after a language split (Starostin, 1989; Starostin, 2000; Embleton, 2000). The basic principles of this method, its applications, and the formulas used in our study are described in the Supplementary Note 1. This glottochronological approach was first used by Starostin with North Caucasus languages (Nikolaev and Starostin 1995). The present study continues this analysis with updated linguistic databases (word lists). The Caucasian word lists used in our study were significantly modified by Mudrak, while the Ossetian word lists were recorded by Ershler and analyzed by Dybo (this study).
Note that both the linguistic and genetic dating methods used in our study provide a most recent (lower) estimate of the population split time (Supplementary Note 1).
RESULTS
Structuring of the Caucasus Y-chromosomal gene pool
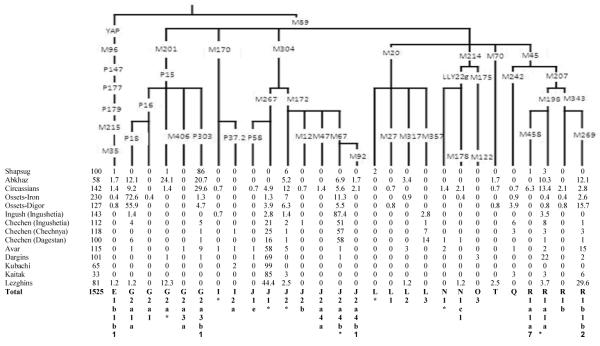
We analyzed 1,525 Y-chromosomal haplotypes from fourteen Caucasus populations. Table 2 presents the haplogroup frequencies, while the Y-STR genotypes are provided in the Supplementary Table 1. We additionally subtyped 121 haplogroup G-M201 samples and 133 haplogroup J-M304 samples from previously-analyzed Near Eastern populations (Supplementary Table 2). Overall, the most frequent haplogroups in the Caucasus were G2a3b1-P303 (12%), G2a1a-P18 (8%), J1*-M267(xP58) (34%), and J2a4b*-M67(xM92) (21%), which together encompassed 73% of the Y chromosomes, while the other 24 haplogroups identified in our study comprise the remaining 27% (Table 2).
Table 2.
Frequencies of Y-chromosomal haplogroups (percent)
|
However, these average frequencies masked the real pattern that became apparent when regional populations were considered (Figure 1). Each of these four haplogroups had its own focus within the Caucasus. More specifically, haplogroup G2a3b1-P303 comprised at least 21% (and up to 86%) of the Y chromosomes in the Shapsug, Abkhaz and Circassians (Figure 1 and Table 2). These populations live in the western part of the Caucasus, and linguistically belong to the Abkhazo-Adyghe language group. The frequency of this haplogroup was below 10% (average 2%) in all other populations investigated from the Caucasus. Similarly, haplogroup G2a1a-P18 comprised at least 56% (and up to 73%) of the Digorians and Ironians (both from the Central Caucasus Iranic linguistic group), while not being found at more than 12% (average 3%) in other populations. Again, haplogroup J2a4b*-M67(xM92) comprised 51-79% of the Y chromosomes in the Ingush and three Chechen populations (North-East Caucasus, Nakh linguistic group), while, in the rest of the Caucasus, its frequency was not higher than 9% (average 3%). Finally, haplogroup J1*-M267(xP58) comprised 44-99% of the Avar, Dargins, Kaitak, Kubachi, and Lezghins (South-East Caucasus, Dagestan linguistic group) but was less than 25% in Nakh populations and less than 5% in the rest of Caucasus.
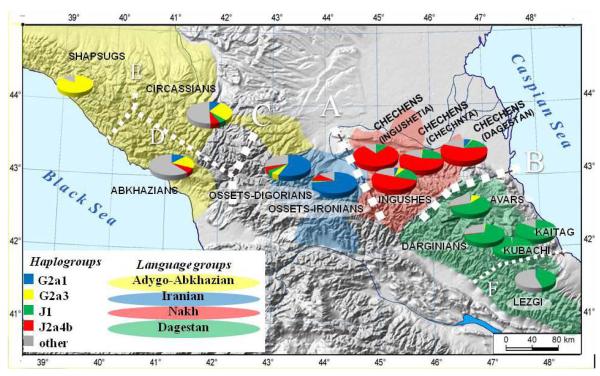
Figure 1. Geographic location, linguistic affiliation and genetic composition of the studied populations.
Each population is designated by a pie chart representing frequencies of the major haplogroups in it. Areas of the linguistic groups of the Caucasus (except for Turkic groups) are shown by semi-transparent color zones. Black dotted lines indicate genetic boundaries identified in the barrier analysis (thick lines – most important boundaries A, B and C; thin lines – other boundaries D, E and F).
A genetic boundary analysis revealed the same pattern (Figure 1). This methodology (Womble, 1951; Rosser et al., 2000) identifies zones of abrupt changes in haplogroup frequencies. The first (most significant) boundary A separated Nakh-Dagestan speaking populations of the east Caucasus from other populations of the region. Boundary B separated the four Nakh-speaking populations from the five Dagestan-speaking ones, while Boundary C separated the Iranian-speaking Ossets (Central Caucasus) from Abkhazo-Adyghe speakers of the west Caucasus. Overall, the first three boundaries divided the Caucasus into four regions, each of which coincided with areas of prevalence of one of the four major Caucasus haplogroups and with areas of different major linguistic groups. The other genetic boundaries D, E and F subdivided Abkhazo-Adyghe speaking populations and separated the Lezghins from populations speaking Dargin languages.
Geography versus Linguistic Diversity
The observed pattern of genetic variation can be interpreted in two ways. On the one hand, it shows excellent correlation with geography, as each of four major haplogroups is prevalent in a specific region of the Caucasus. On the other hand, this pattern also closely fits the classification of Caucasus languages (with no exceptions).
To test these patterns, we computed three matrices of pairwise distances between all studied populations: genetic (from haplogroup frequencies), geographic (in kilometers) and linguistic (percentage of words in common). The Mantel test results showed a significant (p ≤ 0.002) correlation both between genetics and language (r = 0.64) and between genetics and geography (r = 0.60) (Table 3). Partial correlations were calculated between genetics and both factors (language or geography) separately, holding the alternative factor constant. Unfortunately, the analysis was complicated by the fact that linguistics and geography in Caucasus are closely linked with each other (r = 0.78). For this reason, genetic distances exhibited insignificant partial correlation with both, although the correlation with linguistics was almost twice as strong (Table 3).
Table 3.
| Distance considered | Correlation coefficient | P-value |
|---|---|---|
| Genetics and language | 0.64 | 0.002 |
| Genetics and geography | 0.60 | 0.001 |
| Genetics and language. geography held constant | 0.34 | 0.120 |
| Genetics and geography. language held constant | 0.21 | 0.180 |
Populations of the North Caucasian linguistic family (and their respective languages) from Table 1 were considered, except for the genetic isolates Shapshug, Kubachi and Kaitak with census size less than 10,000 persons, as genetic drift may have caused substantial random fluctuation of the haplogroups frequencies in these populations.
Three Chechen populations speaking the same Chechen language were pooled for this analysis. Ossets were not considered because they belong to the Indo-European linguistic family, and linguistic distances cannot be estimated between such divergent languages by this method.
AMOVA was used to further investigate which factor might be the major driving force behind this degree of differentiation (Table 4). When populations were grouped geographically, the proportion of variation in haplogroup frequencies between geographic groups was 0.146. Linguistic classification of the same populations provided nearly two times the extent of variation between linguistic groups (0.268). Therefore, linguistics explained a larger part of Y-chromosomal variation in the Caucasus.
Table 4.
AMOVA results: linguistic versus geographic grouping of populations.
| Linguistic groupinga |
Geographic groupinga |
|
|---|---|---|
| Variation among groupsb | 0.268 * | 0.146 * |
| Variation among populations within groups |
0.099 * | 0.235 * |
| Variation within populations | 0.633 * | 0.619 * |
The linguistic and geographic affiliation of each population is listed in Table 1
P-value <0.001
All populations from Table 1 were used. Thus, AMOVA used a larger data set than the Mantel test because it was possible to use information from the Indo-European speaking Ossetian populations, which were too linguistically distant to obtain lexicostatistical distances for the Mantel test.
These analyses indicated that linguistic diversity is at least as important as geography in shaping the Y-chromosomal landscape, and suggested that the pronounced genetic structure of the Caucasus might have evolved in parallel with the diversification of the North Caucasus languages.
Caucasus in a Eurasian Context
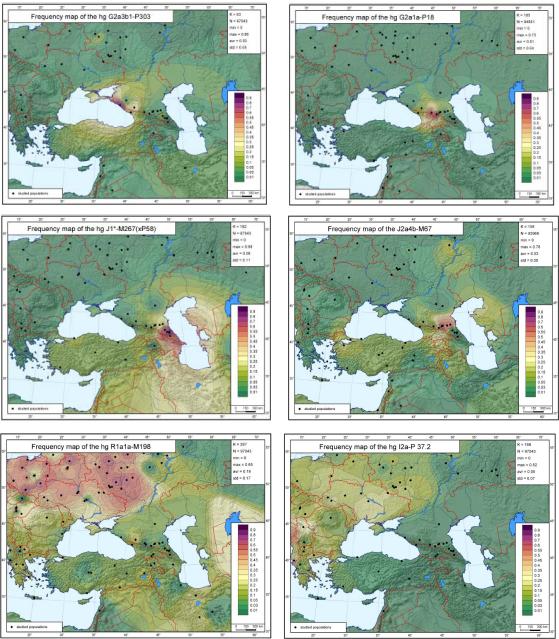
Figure 2 compares the Y-chromosomal pool of the Caucasus with its neighboring regions (Balkans, South-East Europe and the Near East) by presenting frequency maps for haplogroups which predominate in any of these regions. Four haplogroups, G2a3b1-P303, G2a1a-P18, J2a4b*-M67(xM92) and J1*-M267(xP58), exhibit their highest documented frequencies in the Caucasus. Haplogroup G2a3b1-P303 predominates in the West Caucasus (Table 2), although we also found it in the Near East (Table 2) and in one Russian population (data not shown), and it has been reported in Western Europeans (www.ysearch.org). The second haplogroup G2a1a-P18 is almost absent outside of the Caucasus (Figure 2), although its ancestral clade, G2a1-P16, is present in the Near East (Cinnioglu et al., 2004; Flores et al., 2005). Similarly, J1*-M267(xP58) was found mainly in the Caucasus (Figure 2) with the ancestral J1-M267 being common in the Near East. The fourth haplogroup J2a4b*-M67(xM92) is prevalent in a region spanning the Near East and the Caucasus (Figure 2). Note that only a few Near Eastern haplogroups are represented in the Caucasus, and other major components of the Near Eastern Y-chromosomal pool (e.g., J2b-M12) are virtually absent there.
Figure 2.
Frequency maps of major Caucasus, Near Eastern and East European haplogroups.
When haplogroups common in Europe were examined, we observed that the typical Balkan haplogroup I2a-P37.2 was virtually absent in the Caucasus (Figure 2). The recently-defined sub-branch R1a1a7-M458 (Underhill et al., 2010) was found among only the Circassians and Shapsug (Table 2). R1a*-M198(xM458) has an average frequency in the Caucasus as low as 5%, but was found in 20% of the Circassians and 22% of the Dargins, two populations that occupy opposite parts of the Caucasus. STR haplotypes from these Circassian and Dargins samples formed distinct clusters in a network (Supplementary Figure 1). Similarly, two different haplotype clusters within R1b1b2-M269 (Supplementary Figure 1) were found in the Lezghins (30%) and in Ossets-Digor (16%). These concentrations of (presumably European) haplogroups R1a*-M198(xM458), R1a1a7-M458 and R1b1b2-M269 found in few locations in the Caucasus might indicate independent migrations from Europe that were too small to make any significant impact on Caucasus populations.
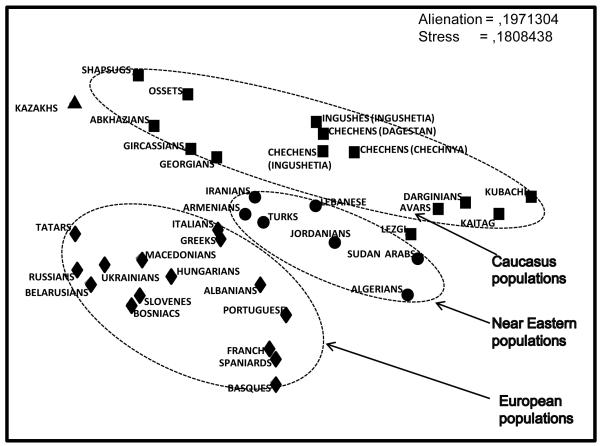
The MDS plot of pairwise genetic distances (Figure 3) showed strong regional clustering separating populations from Europe, the Near East, and the Caucasus, with samples from the Caucasus grouping closer to the Near East samples than to the European ones. Of those three clusters, the Caucasus appeared to be the most diverse, with distinguishable subgroup variation within it. The first subgroup included Dagestan speakers (Avar, Dargins, Kubachi, Kaitak), the second Nakh speakers (three Chechen populations and the Ingush), and the third Abkhazo-Adyghe speakers (Abkhaz, Circassians, Shapsug). Ossets also joined this cluster because we combined their predominant haplogroup G2a1a-P18 with Abkhazo-Adyghe predominant haplogroup G2a3b1-P303 to achieve compatibility with the less phylogenetically resolved European and Near Eastern data. This plot illustrates both the common Near Eastern background of all Caucasus populations and the pronounced intra-Caucasus genetic differentiation into groupings that corresponded well with their linguistic affiliation.
Figure 3. MDS plot depicting genetic relationships between Caucasus, Near Eastern and European populations.
The plot is based on Nei’s pairwise genetic distances calculated from frequencies of thirteen Y chromosomal haplogroups (C, E, G, I, J1, J2, L, N1c, O, R1a1, R1b1, Q, other) in populations of North Caucasus (this study), Transcaucasus (Georgians, Battaglia et al., 2009), Near East (this study; Cinnioglu et al., 2004; Flores et al., 2005), and some other European, African and Asian populations (data from Y-base, compiled in our lab from published sources). Caucasus populations are shown by squares, Near Eastern populations by circles and European populations by diamonds.
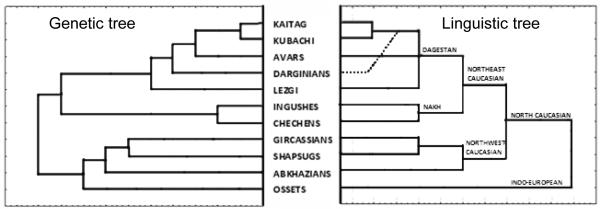
More insights into the relationships between Caucasus populations were obtained from a tree based on haplogroup frequencies (Figure 4, left). Another tree (Figure 4, right), representing the linguistic classification, had the same topology except for the Dargins, who joined the Kubachi/Kaitak cluster before the Avar did. The Indo-European-speaking Ossets were outliers in the Caucasus linguistic tree, and the genetic tree also placed them separately, with slight similarity to the Abkhaz. Generally, the tree based on genetic distances mirrored the linguistic tree in its overall pattern and in most details.
Figure 4. Comparison of the genetic and linguistic trees of North Caucasus populations.
The genetic tree was constructed from frequencies of 28 Y-chromosomal haplogroups in North Caucasus populations (data from Table 2). Populations speaking the same language (three Chechen populations and two Ossetian ones) were pooled to make the genetic dataset compatible with the linguistic classification. The weighted pair-group method was used as a clustering algorithm. The linguistic tree represents the classification of the North Caucasian languages from classical work (Ruhlen, 1987). Kubachi and Kaitak (languages of small populations) were not listed in Ruhlen’s classification, but most linguists agree that they are most related to the Dargin language.
Haplotype networks and age estimates
To analyze this parallelism further, we constructed phylogenetic networks for Caucasus haplogroups (Figure 5, Supplementary Figure 1). While all of the results presented above were obtained using only haplogroup frequencies, in this and the following section we analyze the variation of STR haplotypes within haplogroups.
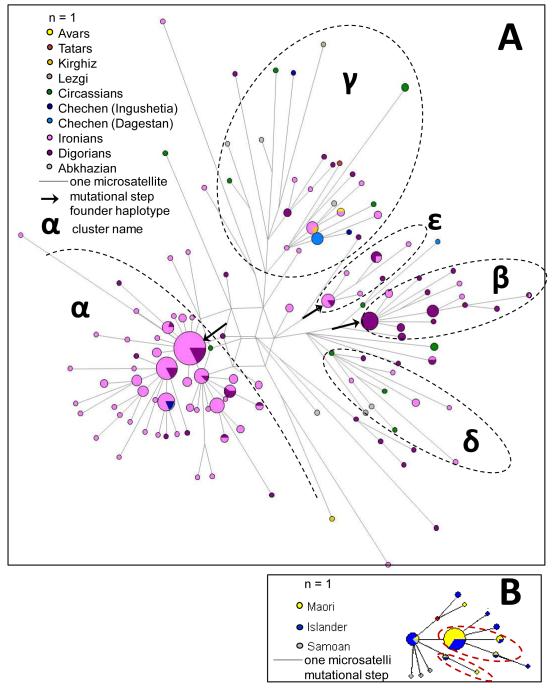
Figure 5. Phylogenetic networks of the haplogroup G2a1a-P18 in the Caucasus and haplogroup C-M208 in Polynesia.
A. Reduced median network of haplogroup G2a1a-P18 was constructed using all available (worldwide) STR haplotypes for this haplogroup, with a reduction threshold r=1.00 based on non-weighted data from 15 STRs (DYS19, DYS389I, DYS389b, DYS390, DYS391, DYS392, DYS393, DYS437, DYS438, DYS439, DYS448, DYS456, DYS458, DYS635, GATA_H4). Black dotted lines designate clusters selected in our study for age estimations.
B. Reduced median network of haplogroup C-M208 in Polynesia (modified from Zhivotovsky et al., 2004). Red dotted lines designate clusters selected by Zhivotovsky et al. (2004) for estimating the evolutionary effective mutation rate.
Haplogroup G2a1a-P18 (Figure 5), which was found almost exclusively in the Caucasus, consisted of distinct branches of STR haplotypes rooted in the central reticulated zone. The larger cluster α includes mainly Ossets-Iron and demonstrated a star-like pattern with a central founder haplotype and a few subfounders. The cluster β comprised many samples of Ossets-Digor, while smaller cluster γ comprised both Ossetian populations. The diverse cluster in the upper part of the network comprised different Caucasus populations, and a few non-Caucasus G2a1a-P18 samples also belonged to this branch.
Generally, haplogroup G2a1a-P18 seemed to have a long history in the Caucasus, being spread across the region and forming many branches. However, two of them (clusters α and β, found in the Ironians and Digorians, respectively) showed in their star-like structure signs of relatively recent expansion. The average number of mutation steps (ρ estimator) was similar for both clusters (1.46 for α and 1.41 for β), indicating that both clusters had expanded at around the same time, possibly because of the same event.
Reduced median networks for other haplogroups (Supplementary Figure 1) revealed similar patterns of branching. Some branches were shared between different Caucasus populations (and often included also Near Eastern samples), while many others were absent from the Near East and, moreover, specific to individual Caucasus populations (e.g. clusters α, β, and γ in Figure 5 are specific to the Ossets).
Applying the formal criteria, we identified 18 population-specific clusters (average specificity 95%). Table 5 lists these clusters with their ages, suggests the population event relevant to each, and indicates the linguistic date of this event (the tree of the North Caucasian languages obtained in our study is presented in Supplementary Figure 2).
Table 5.
Genetic and linguistic dates of the populations splits.
| Cluster | Specific to population |
STRsa | NSb | NH c | S d | ρ e ± σ f | AgeG g | AgeE h | AgeB i | Linguistic date |
Population event |
|---|---|---|---|---|---|---|---|---|---|---|---|
| P303-β | Shapsug and Circassians |
15 | 12 | 7 | 100 | 1.5±0.5 | 1400±500 | 4300±1400 | MIN(1.1-4.8) MAX(1.8-7.8) | 3600 | Separation of Shapsug- Circassians branch from Abkhaz |
|
| |||||||||||
| P303-α | Shapsug | 15 | 27 | 8 | 92.6 | 0.55±0.25 | 500±200 | 1600±700 | MIN(1.0-2.6) MAX(1.4-3.9) | 800 | Separation of Shapsug from Circassians |
|
| |||||||||||
| P18-α | Ossets-Iron | 15 | 211 | 49 | 88.6 | 1.46±0.48 | 1400±500 | 4200±1400 | MIN(2.5-5.2) MAX(2.9-6.7) | 1300 | Split of Ossets into Iron and Digor |
| P18-β | Ossets-Digor | 15 | 28 | 12 | 85.7 | 1.41±0.48 | 1300±500 | 4100±1400 | MIN(1.1-2.5) MAX(1.4-3.3) | 1300 | |
| R1b1b2-β | Ossets-Digor | 17 | 24 | 12 | 100 | 0.91±0.31 | 800±300 | 2300±800 | MIN(1.1-3.0) MAX(1.9-5.3) | 1300 | |
|
| |||||||||||
| P18- γ | Ossets-Iron and Digor |
15 | 10 | 5 | 100 | 1±0.52 | 1000±500 | 2900±1500 | - | - | - |
|
| |||||||||||
| M67-β | Chechen and Ingush |
14 | 45 | 12 | 93 | 1.56±0.81 | 1600±800 | 4800±2500 | MIN(1.0-4.0) MAX(1.8-6.7) | 5600 | Separation of Nakh populations from Dagestan ones |
| M67-γ | Chechen and Ingush |
14 | 81 | 22 | 96 | 1.84±0.69 | 1900±700 | 5700±2100 | MIN(0.9-2.5) MAX(1.5-4.1) | 5600 | |
|
| |||||||||||
| M67- α | Ingush | 14 | 53 | 9 | 81 | 1.96±1,02 | 2000±1000 | 6100±3200 | MIN(1.4-3.2) MAX(1.9-4.2) | 1400 | Split of Nakh branch into Chechen and Ingush |
| M67-δ | Chechen | 14 | 22 | 3 | 100 | 0.14±0.1 | 100±100 | 400±300 | - | 1400 | |
| L3 | Chechen | 17 | 24 | 13 | 95.8 | 1.13±0.42 | 900±400 | 2900±1100 | MIN(0.9-3.3) MAX(11.9-20.4) | 1400 | |
| Q-α | Chechen | 17 | 10 | 6 | 100 | 2.18±1.02 | 1800±900 | 5600±2600 | MIN(0.6-2.2) MAX(1.2-4.6) | 1400 | |
|
| |||||||||||
| M267(xP58)α | Dargins. Kubachi |
15 | 16 | 6 | 100 | 1.06±0.5 | 1000±500 | 3100±1400 | MIN(0.4-1.7) MAX(0.7-3.0) | 3400 | Separation of Dargins. Kubachi and Kaitak from other Dagestan populations |
| M267(xP58)β | Dargins Kubachi |
15 | 15 | 8 | 93.3 | 1.44±0.63 | 1400±600 | 4200±1800 | MIN(0.7-2.3) MAX(1.2-3.3) | 3400 | |
| M267(xP58)γ | Dargins. Kubachi. Kaitak |
15 | 11 | 6 | 100 | 1.09±0.68 | 1000±600 | 3200±2000 | MIN(0.5-1.9) MAX(1.0-3.1) | 3400 | |
|
| |||||||||||
| R1a1a*-α | Dargins | 17 | 13 | 5 | 100 | 1.77±0.99 | 1500±800 | 4500±2500 | MIN(0.4-1.4) MAX(0.6-2.7) | 1900 | Separation of Kubachi from Dargins |
|
| |||||||||||
| R1b1b2-α | Lezghins | 17 | 21 | 9 | 100 | 2.38±0.89 | 2000±700 | 6100±2300 | MIN(1.0-2.7) MAX(1.3-3.7) | 4300 | Separation of Lezghins from Dargins populations |
| P15*-α | Lezghins | 15 | 11 | 8 | 90.9 | 2.45±0.76 | 2300±700 | 7100±2200 | MIN(1.2-3.7) MAX(1.7-5.6) | 4300 | |
Number of STRs used for the network
Number of samples in cluster
Number of haplotypes in cluster
Specificity of the clusters (proportion of samples from the indicated population among all samples within the cluster, in percent)
Average distance to the founder haplotype (Forster et al., 1996).
Estimator for the variance (Saillard et al., 2000).
Age of the cluster, obtained from the ρ estimator using the “genealogical” mutation rate (y BP).
Age of the cluster, obtained from the ρ estimator using the “evolutionary” mutation rate (y BP).
95% CI of age of the NETWORK-identified STR haplotype cluster within the population obtained in the BATWING analysis using the “genealogical rate” (ky BP).
A tree of population splits
Because of the controversy between “evolutionary” and “genealogical” mutation rates, we set out to reconstruct the population history of the Caucasus in two phases. The first was based solely on genetic evidence without consideration of any mutation rates, while the second converted genetic diversity into time using both rates and then compared them with the linguistic times.
In phase 1, we grouped populations according to the predominant haplogroup, which yielded four branches (Supplementary Figure 3). To explore the intra-branch relationships, we examined the population-specific STR-clusters (Table 5). Cluster P303-β was shared between Shapsug and Circassians, while no cluster linked any of these populations with Abkhaz. We concluded that Abkhaz separated first, while Shapsug and Circassians maintained a shared ancestry for a longer period, during which the P303-β cluster originated. The cluster P303-α, which is present in Shapsug but absent in Circassians, marks the next split on the population tree. In terms of mutation steps, the first split (Abkhaz vs Shapsug-Circassians) occurred 1.5 “mutations before present”, while the second split (Shapsug vs Circassians) took place 0.6 “mutations before present” (Table 5).
Applying the same methodology to another three branches of populations led to a tree of population splits (Supplementary Figure 3). This tree is based solely on genetic data and yet shows good agreement with the topology of both linguistic trees: the classical way of grouping North Caucasian languages (Figure 4, right) and the quantitative lexico-statistical tree (Supplementary Figure 2). Up to this point, we have avoided using any mutation rate. As a result, we compare the topology of the trees (sequence of splitting events) without reference to a time scale.
In phase 2, to introduce time estimates, we used both “genealogical” and “evolutionary” mutation rates. The ρ estimator using the genealogical rate provided a good fit between genetics and linguistics, as the genetic dates were similar to, or younger than, the linguistic dates. (Because clusters can expand at any time after the split, they are expected to be younger than the respective languages; however, they should not be older, as described in Supplementary Note 1. Estimates based on the “evolutionary” mutation rate were too old to be in agreement with the linguistic dates (Table 5).
BATWING computations of the ages of the same clusters based on genealogical rates showed similar results to those indicated by the linguistic analysis. The age for the four major haplogroups in individual populations obtained by using SD estimator (Supplementary Table 3) are close to the Neolithic epoch, and might be interpreted as signs of population expansion due to the shift to a farming economy.
The BATWING tree of population splits (Supplementary Figure 4) was based on all STR data from Supplementary Table 1 disregarding the haplotype clusters to which they belong. This tree therefore provides an independent test of the other genetic trees presented in this study (the tree on the left of Figure 4 is based on the haplogroup frequencies, while trees in Figure 6 and Supplementary Figure 3 are based on the ages of haplotype clusters). This tree again showed a striking resemblance to the linguistic tree: one observes an initial split into west and east Caucasus populations, and then the separation of Abkhaz from Circassian-Shapsug on the western branch and Nakh populations from Dagestan ones on the eastern branch; disagreements could be found only within the Dagestan group. Ossets (linguistic outliers) showed a slight similarity to Abkhaz, as they did on the haplogroup-based tree, as well (Figure 4 left). When the topology of this tree is similar to the linguistic one, the BATWING dates were on average 1.5 times younger. If the evolutionary mutation rate were applied (data not shown), the topology of the BATWING tree would remain the same but the dates would become on average 1.5 times older than corresponding linguistic dates.
Table 6.
Endogamy levelsa for Caucasus and some European populations.
| Population | Sample size | Level of endogamy (%) |
Random inbreeding e |
Method for estimating inbreeding |
Reference |
|---|---|---|---|---|---|
| NТ | fr 102 102
|
||||
| Shapsug | 5,928 | 95.2 b | 2.86 | Isonymy |
Balanovska, E.,V. et al., 2000; Pocheshkhova, E., A. 2008 |
| Abkhaz | 253 | 98 c | 3.13 | Isonymy | Pocheshkhova, E., A. 2008 |
| Circassians | 4,438 | - | 0.61 | Isonymy | Pocheshkhova, E., A. 2008 |
| Kubachi | 182 | 99.0 d | 1.21 | Demography | Bulaeva, K., B. et al., 2004 |
| Dargins | 350 | 92.2 d | 1.19 | Demography | Bulaeva, K., B. et al., 2004 |
| Avar | 299 | 86.7 d | 1.03 | Demography | Bulaeva, K., B. et al.,1990 |
| Botlikh | 248 | 75.6 d | 0.50 | Demography | Bulaeva, K., B. et al.,1990 |
| Andi | 171 | 87.0 d | 1.20 | Demography | Bulaeva, K., B. et al.,1990 |
| Tindal | 374 | 91.0 d | 1.12 | Demography | Bulaeva, K., B. et al.,1990 |
| Lak | 349 | 81.3 d | 0.011 | Demography | Bulaeva, K., B. et al., 1990 |
| Lezghins | 175 | - | 0.99 | Demography | Bulaeva, K., B. et al., 2004 |
| Mormon | 625 | - | 0.19 | Demography | Vogel, F., Motulsky, A.G. 1986 |
| Kuban Cossacks |
16,056 | - | 0.06 | Isonymy | Pocheshkhova, E., A. 2008 |
| French | 530,000 | - | 0.02 | Demography | Vogel and Motulsky, A.G. 1986 |
| Irish | 190,547 | - | 0.02 | Demography | Vogel and Motulsky, A.G. 1986 |
| Italians | 1,646,612 | - | 0.07 | Demography | Vogel and Motulsky, A.G. 1986 |
| Switzerland (4 mountain villages) |
538 | - | 0.51 | Demography | Vogel and Motulsky, A.G. 1986 |
The table summarizes traditional marriage practices in Caucasus. These data indicate high ethnic endogamy and concomitantly high levels of inbreeding as a consequence.
The proportion of endogamous marriages was estimated for ethnic group
The proportion of endogamous marriages was estimated for administrative districts (intraethnic level)
The proportion of endogamous marriages was estimated for villages (local level). Most estimates were obtained for villages because inter-ethnic marriages for these groups are typically less than 1%.
The random inbreeding fr was estimated as the proportion of isonymy marriages expected in the panmictic population (Crow and Mange, 1965)
DISCUSSION
Origin and structuring of the North Caucasus paternal pool
Four haplogroups are predominant in the Caucasus (Table 2), and each of them has its own domain (recognizable geographically and also linguistically), where it represents the lion’s share of the regional gene pool. In all other domains, the given haplogroup is infrequent or absent. The robustness of this conclusion is enhanced by the fact that each domain is occupied by more than one population in our data set whose characteristic haplogroup is prevalent in each population of the domain (Table 1). This pronounced structuring of the Y-chromosomal pool of the Caucasus has not previously been reported. In the study of Nasidze et al. (2004), AMOVA computation revealed a lack of correlation with language, while the correlation with geography did not reach the significance threshold. The increased phylogenetic resolution and large sample sizes used in our study were necessary to reveal the links between haplogroups, regions and language groups (Figure 1). Methodologically, the analysis of correlations between geography, language and genetics (Table 3) parallels an earlier study performed with European populations (Rosser et al., 2000), where geography was shown to be the leading factor in Europe. In our analysis, the correlation was much stronger (maximum value 0.64) than in the European study (0.39).
Strikingly, language rather than geography tended to have a larger influence on the genetic structuring in the Caucasus (Tables 3 and 4). Language and geography are also closely linked with each other, probably because of the mountainous nature of the Caucasus region where languages are often restricted to a few valleys. In this context, the slightly higher dependence of genetic structure on language could be explained by marriage and individual migration practices, linking linguistically similar populations in preference. For example, Circassians, who are geographically situated between Adyghes and Ossets, might receive more gene flow from Adyghes, who speak a similar language, than from Ossets who differ in their language and culture.
Ossets, who speak an Indo-European language, find their place among populations of the North Caucasus language family. This genetic association is consistent with the physical anthropological evidence (Abramova, 1989; Melyukova, 1989) that Ossets are mainly descendants of indigenous Caucasus populations, who were assimilated by Alans and received from them the present language. Little is known about the language that these populations initially spoke. Note that, on the genetic trees, Ossets join the western (Abkhaz-Adyghe) branch of the North Caucasus family.
Although occupying a boundary position between Europe and the Near East, all four major Caucasus haplogroups show signs of a Near Eastern rather than European origin (Figure 2, Supplementary Figure 1). These four haplogroups reach their maximum (worldwide) frequencies in the Caucasus (Table 2, Figure 2). They are either shared with Near East populations (G2a3b1-P303 and J2a4b*-M67(xM92)) or have ancestral lineages present there (G2a1*-P16(xP18) and J1*-M267(xP58)). Typical European haplogroups are very rare (I2a-P37.2) or limited to specific populations (R1a1a-M198; R1b1b2-M269) in the Caucasus.
This pattern suggests unidirectional gene flow from the Near East towards the Caucasus, which could have occurred during the initial Paleolithic settlement or the subsequent Neolithic spread of farming. Archaeological data do not indicate a Near Eastern influence on the Neolithic cultures in the North Caucasus (Bader and Tseretely, 1989; Bzhania, 1996; Masson et al., 1994), while Neolithization in the Transcaucasus was part of a Neolithic expansion that perhaps paralleled those occurring in Europe (Balaresque et al. 2010) and North Africa (Arredi et al. 2004). However, the current genetic evidence does not allow us to distinguish between Paleolithic and Neolithic models in shaping the genetic landscape of the North Caucasus.
All of these genetic findings are based solely on Y-chromosomal data. This choice was prompted by the high inter-population variation in the data set (and therefore the best detection of the differences) compared with mtDNA and autosomal markers. However, one may wonder if the pattern of the entire gene pool is different from its Y chromosomal subset. In the context of this study, languages are typically learned from the maternal side (we say “mother tongue”; Beauchemin et al., 2010). Thus the observed similarity between the distributions of languages and genes might become even more evident if full-genome data, incorporating maternally-inherited information as well, become available; this possibility may be explored in future studies.
Challenges of genetic dating
To estimate the ages of population splits, we employed both “evolutionary” and ”genealogical” mutation rates for calibration and used four different methods, namely the ρ estimator, BATWING dating of the clusters and the population splits, and ASD microsatellite variation.
The reliability of the ρ estimator has been explored by Cox (2008) using simulations under a number of demographic models. He found that the mean age is biased only slightly, but the confidence intervals might not contain the true value in 34% cases for the simplest model (constant effective population size, Ne = 1000). In some demographic conditions, the error rate increases, particularly when samples sizes are below 25, or when Ne is large, unstable (bottlenecks) or growing. The Ne estimates available for the Caucasus populations are small (Ne = 187 on average; Pocheshkhova, 2008), which might indicate that the error rate should be low. However, Caucasus populations did grow and bottlenecks could not be excluded. In fact, the most pronounced demographic feature of the Caucasus populations is their high degree of subdivision; fortunately, “error rates of molecular dating with the ρ statistic are unaffected by simple population subdivision” (Cox, 2008). Therefore, we might expect ~34% of our clusters (Table 5, Figure 6) to have actual ages falling outside the indicated confidence intervals. This factor, which randomly affects only one-third of the clusters, would not eliminate the overall agreement with linguistics seen from the Figure 6, although it highlights the fact that genetic dates for each particular branch should be taken with caution.
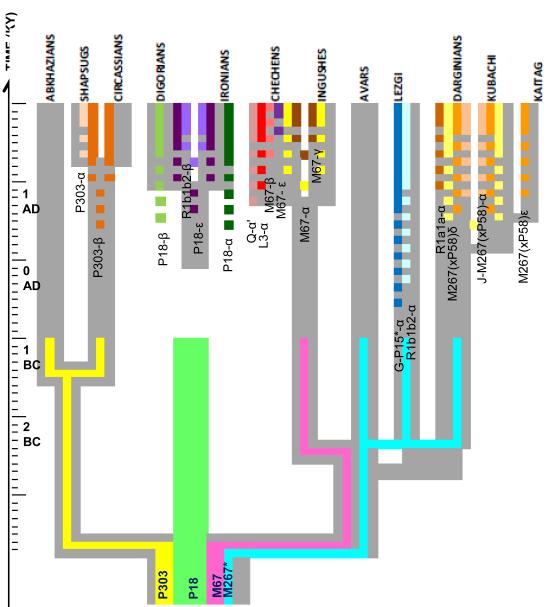
Figure 6. Model of the evolution of Caucasus populations combining genetic and linguistic evidence.
The grey background outlines the linguistic tree, obtained by lexicostatistical method. Each colored line near the tips of the tree marks a haplotype cluster that is specific to a given population. If the cluster is shared between two populations, then both populations carry this color on their branches. Standard errors of a cluster’s age are shown by dotted colored lines. Each colored line near the root of the tree marks one of four major haplogroups. These lines stop 3,300 years BP. The root of the population tree indicates an initial migration from the Near East carrying four major haplogroups. This proto-population then separated into the West Caucasus, proto-Ossets, Nakh and Dagestan branches, differing by language and predominant haplogroup. The subsequent evolution (occurring independently in each of these four groups) consisted in the diversification of their languages and emergence of branch-specific or population-specific haplotype clusters.
We found that “evolutionary” estimates of most clusters fall far outside the range of the respective linguistic dates, while “genealogical” estimates gave a good fit with the linguistic dates. At least two population events in the Caucasus are documented archaeologically, which allows additional comparison with these “historical” dates. In both cases, the historical (archaeological) date is similar to a genetic estimate based on the “genealogical” mutation rate (Supplementary Note 2). In this regard, a study of the link between Y-chromosomes and British surnames working with time intervals close to those analyzed here obtained a mutation rate of 1.5 × 10−3 (King and Jobling, 2009). This rate is similar to the “genealogical” rather than the “evolutionary” rate, and provided good agreement with the historical dates for the surname ages.
The “evolutionary” rate (Zhivotovsky et al., 2004) was calibrated using two contrasting populations (Maori and Roma). The fact that, in the Caucasus, the genealogical rate provides a better fit with history and linguistics might be partly explained by the dependence of the estimated intra-lineage variance (and therefore age) on the way that clusters are selected in the network. We selected larger clusters, containing 11 haplotypes on average. However, when the evolutionary calibration was performed (Zhivotovsky et al., 2004), large data sets were not available and clusters contained only 2-4 haplotypes. For example, the Zhivotovsky et al. (2004) study chose two founders in the Polynesian network (Figure 5B), while, in our study, we considered similar topologies as a single cluster (see cluster γ in Figure 5A for comparison). These subclusters (Figure 5B) might be justified in the case of the peopling of New Zealand because they could originate in the homeland before the migration to New Zealand. In our case, however, we considered clusters within the networks because we were interested in the whole history of the clusters that had arisen in situ within the Caucasus. To avoid arbitrarily identifying the clusters, we followed a set of formal rules, as described above.
It should be mentioned here that, for the BATWING tree (which does not require identifying the clusters), applying the genealogical rate underestimates the dates, while applying evolutionary rates overestimates the dates. These comparisons were made with 14 linguistic dates, but more sophisticated modeling and calibrations in other regions are needed to find the most appropriate way to incorporate mutation rate estimates into population-genetic applications. Our study shows that the results could be affected by the method of identifying the clusters and particularly by the chosen methods of dating (ρ, BATWING, SD).
CONCLUSION
Combining genetic and linguistic findings, we now propose a model of the evolution of the Caucasus populations. The final tree (Figure 6) was obtained by merging the genetic clusters with the background linguistic tree. We conclude that the Caucasus gene pool originated from a subset of the Near Eastern pool due to an Upper Paleolithic (or Neolithic) migration, followed by significant genetic drift, probably due to isolation in the extremely mountainous landscape. This process would result in the loss of some haplogroups and the increased frequency of others. The Caucasus meta-population underwent a series of population (and language) splits. Each population (linguistic group) ended up with one major haplogroup from the original Caucasus genetic package, while other haplogroups became rare or absent in it. The small isolated population of the Kubachi, in which haplogroup J1*-M267(xP58) became virtually fixed (99%, Table 2), exemplifies the influence of genetic drift there. During population differentiation, haplotype clusters within haplogroups emerged and expanded, often becoming population-specific. The older clusters became characteristic of groups of populations. Many younger clusters were specific to individual populations (typically speaking different languages).
We note that the method of inferring the topology of the genetic tree of the Caucasus populations does not require the inference of any mutation rates, and the result is strikingly concordant with the topology of the linguistic tree. Mutation rates are required only for adding a time scale to both trees. Based on the topologies of trees generated from both the genetic and linguistic data, the inference of the parallel evolution of genes and languages in Caucasus is supported, despite controversies about the mutation rates. This study of Caucasus Y-chromosomal variation demonstrates that genetic and linguistic diversification were two parallel processes or, perhaps more precisely, two sides of the same process of evolution of the Caucasus meta-population over hundreds of generations.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the people from the Caucasus populations who provided their DNA for the present analysis, Andrey Pshenichnov and Roman Sychev for help in compiling Y-base, Vadim Urasin for collecting STR data from DNA-genealogical websites, Valery Zaporozhchenko for a pilot study of the STR variation of haplogroup G-M201 in the West Caucasus, Kazima B. Bulaeva for consultation about endogamy levels in the East Caucasus, Vladimir Kharkov for help with first version of the phylogenetic network, Matt Kaplan for replication of 20 samples as part of the quality control procedures, Janet Ziegle for technical assistance and two anonymous reviewers for helpful comments. The first author thanks Richard Villems for instruction in phylogenetic methods and helpful discussions.
FUNDING This work received primary support from the Genographic Project and additional support from the Russian Foundation of Basic Research (grants 10-07-00515, 10-06-00451, 10-04-01603). CTS was supported by The Wellcome Trust.
Footnotes
Genographic Consortium members: Syama Adhikarla (Madurai Kamaraj University, Madurai, Tamil Nadu, India), Christina J. Adler (University of Adelaide, South Australia, Australia), Danielle A. Badro (Lebanese American University, Chouran, Beirut, Lebanon), Jaume Bertranpetit (Universitat Pompeu Fabra, Barcelona, Spain), Andrew C. Clarke (University of Otago, Dunedin, New Zealand), David Comas (Universitat Pompeu Fabra, Barcelona, Spain), Alan Cooper (University of Adelaide, South Australia, Australia), Clio S. I. Der Sarkissian (University of Adelaide, South Australia, Australia), Matthew C. Dulik (University of Pennsylvania, Philadelphia, Pennsylvania, United States), Christoff J. Erasmus (National Health Laboratory Service, Johannesburg, South Africa), Jill B. Gaieski (University of Pennsylvania, Philadelphia, Pennsylvania, United States), ArunKumar GaneshPrasad (Madurai Kamaraj University, Madurai, Tamil Nadu, India), Angela Hobbs (National Health Laboratory Service, Johannesburg, South Africa), Asif Javed (IBM, Yorktown Heights, New York, United States), Li Jin (Fudan University, Shanghai, China), Matthew E. Kaplan (University of Arizona, Tucson, Arizona, United States), Shilin Li (Fudan University, Shanghai, China), Begoña Martínez-Cruz (Universitat Pompeu Fabra, Barcelona, Spain), Elizabeth A. Matisoo-Smith (University of Otago, Dunedin, New Zealand), Marta Melé (Universitat Pompeu Fabra, Barcelona, Spain), Nirav C. Merchant (University of Arizona, Tucson, Arizona, United States), R. John Mitchell (La Trobe University, Melbourne, Victoria, Australia), Amanda C. Owings (University of Pennsylvania, Philadelphia, Pennsylvania, United States), Laxmi Parida (IBM, Yorktown Heights, New York, United States), Ramasamy Pitchappan (Madurai Kamaraj University, Madurai, Tamil Nadu, India), Lluis Quintana-Murci (Institut Pasteur, Paris, France), Daniela R. Lacerda (Universidade Federal de Minas Gerais, Belo Horizonte, Minas Gerais, Brazil), Ajay K. Royyuru (IBM, Yorktown Heights, New York, United States), Fabrício R. Santos (Universidade Federal de Minas Gerais, Belo Horizonte, Minas Gerais, Brazil), Himla Soodyall (National Health Laboratory Service, Johannesburg, South Africa), Pandikumar Swamikrishnan (IBM, Somers, New York, United States), Kavitha Valampuri John (Madurai Kamaraj University, Madurai, Tamil Nadu, India), Arun Varatharajan Santhakumari (Madurai Kamaraj University, Madurai, Tamil Nadu, India), Pedro Paulo Vieira (Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil), Janet S. Ziegle (Applied Biosystems, Foster City, California, United States).
LITERATURE CITED
- 1.Abdushelishvili MG. Antropology of the ancient and contemporary population of Georgia. Metsniereba; Tbilisi: 1964. [Google Scholar]
- 2.Abramova MP. The Central Caucasus in the Sarmatian epoch. In: Rybakov BA, editor. The steppes of the European part of the USSR in the Scythian-Sarmatian time. Nauka; Moscow: 1989. pp. 268–281. (Series Archaeology of the USSR). [Google Scholar]
- 3.Ageeva RA. Which tribe we are? Ethnic groups of Russia: ethnonims and fortunes. Ethnolinguistic dictionary. Academia Press; Moscow: 2000. [Google Scholar]
- 4.Alexeev VP. The origin of Caucasus peoples. Mysl; Moscow: 1974. [Google Scholar]
- 5.Alexeev ME. Languages of the world: Caucasus languages. Academia; Moscow: 1999. Nakh-Dagestan languages; pp. 156–165. [Google Scholar]
- 6.Arredi B, Poloni ES, Paracchini S, et al. A predominantly Neolithic origin for Y-chromosomal DNA variation in North Africa. Am. J. Hum. Genet. 2004;75:338–345. doi: 10.1086/423147. (9 co-authors) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bader NO, Tsereteli LD. Mesolithic in the Caucasus. In: Rybakov BA, editor. Mesolithic of the USSR. Nauka; Moscow: 1989. pp. 93–105. (Series Archaeology of the USSR). [Google Scholar]
- 8.Balanovska EV, Pocheshkhova EA, Balanovsky OP, et al. Gene-geographic analysis of a subdivided population. II Geography of random inbreeding based on surname frequencies in Adygs. Russ. J. Genet. 2000;36:1126–1139. (4 co-authors) [PubMed] [Google Scholar]
- 9.Balanovsky O, Rootsi S, Pshenichnov A, et al. Two sources of the Russian patrilineal heritage in their Eurasian context. Am. J. Hum. Genet. 2008;82:236–250. doi: 10.1016/j.ajhg.2007.09.019. (11 co-authors) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balaresque P, Bowden GR, Adams SM, et al. A predominantly Neolithic origin for European paternal lineages. PLoS Biol. 2010;8:1–9. doi: 10.1371/journal.pbio.1000285. (16 co-authors) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bandelt HJ, Forster P, Sykes BC, et al. Mitochondrial portraits of human populations using median networks. Genetics. 1995;141:743–753. doi: 10.1093/genetics/141.2.743. (4 co-authors) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Battaglia V, Fornarino S, Al-Zahery N, et al. Y-chromosomal evidence of the cultural diffusion of agriculture in Southeast Europe. Eur. J. Hum. Genet. 2009;17:820–830. doi: 10.1038/ejhg.2008.249. (18 co-authors) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beauchemin M, González-Frankenberger B, Tremblay J, Vannasing P, Martínez-Montes E, Belin P, Béland R, Francoeur D, Carceller AM, Wallois F, Lassonde M. Mother and Stranger: An Electrophysiological Study of Voice Processing in Newborns. Cereb. Cortex. 2010 Dec 13; doi: 10.1093/cercor/bhq242. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 14.Betrozov R. Origin and ethnocultural contacts of Adyghes. Nart; Nalchik: 1991. [Google Scholar]
- 15.Blazhek V, Novotna P. Retoromanske jazyky: prehled a klasifikace. Linguistica Brunensia. Sbornik praci filozoficke fakulty brnenske univerzity A. 2008;56:15–32. [Google Scholar]
- 16.Bokarev EA. Comparative-historical phonetics of the East Caucasian languages. Nauka; Moscow: 1981. [Google Scholar]
- 17.Bulaeva KB, Isaichev SA, Pavlova TA. Population-genetics approach to the genetics of human behaviour. Biomed Sci. 1990;1:417–424. [PubMed] [Google Scholar]
- 18.Bulaeva KB, Jorde L, Ostler C, et al. STR polymorphism in populations of indigenous Daghestan ethnic groups. Genetika. 2004;40:691–703. (6 co-authors) [PubMed] [Google Scholar]
- 19.Bulaeva KB, Jorde L, Watkins S, et al. Ethnogenomic diversity of Caucasus, Daghestan. Am. J. Hum. Biol. 2006;18:610–620. doi: 10.1002/ajhb.20531. (9 co-authors) [DOI] [PubMed] [Google Scholar]
- 20.Bzhania VV. Caucasus. In: Rybakov BA, editor. Neolithic of the Northern Eurasia. Nauka; Moscow: 1996. pp. 73–86. (Series Archaeology of the USSR). [Google Scholar]
- 21.Caciagli L, Bulayeva K, Bulayev O, et al. The key role of patrilineal inheritance in shaping the genetic variation of Dagestan highlanders. J. Hum. Genet. 2009;54:689–694. doi: 10.1038/jhg.2009.94. (8 co-authors) [DOI] [PubMed] [Google Scholar]
- 22.Chirikba VA. Common West Caucasian: The Reconstruction of its Phonological System and Parts of its Lexicon and Morphology. CNWS Publications; Leiden: 1996. [Google Scholar]
- 23.Cinnioglu C, King R, Kivisild T, et al. Excavating Y-chromosome haplotype strata in Anatolia. Hum. Genet. 2004;114:127–148. doi: 10.1007/s00439-003-1031-4. (15 co-authors) [DOI] [PubMed] [Google Scholar]
- 24.Comrie B. The World’s Major Languages. Oxford University Press; New York: 1987. [Google Scholar]
- 25.Cox MP. Accuracy of molecular dating with the rho statistic: deviations from coalescent expectations under a range of demographic models. Hum Biol. 2008;80:335–357. doi: 10.3378/1534-6617-80.4.335. [DOI] [PubMed] [Google Scholar]
- 26.Crow JF, Mange AP. Measurement of inbreeding from frequency of marriages between person of the same surname. Eug. Quart. 1965;12:199–203. doi: 10.1080/19485565.1965.9987630. [DOI] [PubMed] [Google Scholar]
- 27.Cruciani F, La Fratta R, Santolamazza P, et al. Phylogeographic analysis of haplogroup E3b (E-M215) Y chromosomes reveals multiple migratory events within and out of Africa. Am. J. Hum. Genet. 2004;74:1014–1022. doi: 10.1086/386294. (19 co-authors) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cruciani F, La Fratta R, Torroni A, et al. Molecular dissection of the Y chromosome haplogroup E-M78 (E3b1a): a posteriori evaluation of a microsatellite-network-based approach through six new biallelic markers. Hum. Mutat. 2006;27:831–832. doi: 10.1002/humu.9445. (5 co-authors) [DOI] [PubMed] [Google Scholar]
- 29.Cruciani F, La Fratta R, Trombetta B, et al. Tracing past human male movements in northern/eastern Africa and western Eurasia: new clues from Y-chromosomal haplogroups E-M78 and J-M12. Mol. Biol. Evol. 2007;24:1300–1311. doi: 10.1093/molbev/msm049. (24 co-authors) [DOI] [PubMed] [Google Scholar]
- 30.Di Giacomo F, Luca F, Popa LO, et al. Y chromosomal haplogroup J as a signature of the post-neolithic colonization of Europe. Hum. Genet. 2004;116:529–532. doi: 10.1007/s00439-004-1168-9. (27 co-authors) [DOI] [PubMed] [Google Scholar]
- 31.Dybo AV. Chronology of Turkic languages and linguistic contacts of early Turkic groups. In: Tenishev ER, Dybo Av. V., editors. Comparative historical grammar of Turkic languages. Proto-Turkic basic language. Nauka; Moscow: 2006. pp. 766–820. [Google Scholar]
- 32.Dybo AV. Supplement: One hundred word lists of Turkic languages with the etymological comments. In: Tenishev ER, Dybo Av.V, editors. Comparative historical grammar of Turkic languages. Proto-Turkic basic language. Nauka; Moscow: 2006. pp. 824–861. [Google Scholar]
- 33.El-Sibai M, Platt DE, Haber M, et al. Geographical structure of the Y-chromosomal genetic landscape of the Levant: a coastal-inland contrast. Ann. Hum. Genet. 2009;73:568–581. doi: 10.1111/j.1469-1809.2009.00538.x. (38 co-authors) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Embleton S. Lexicostatistics. Glottochronology: from Swadesh to Sankoff to Starostin to future horizons. In: Renfrew C, McMahon A, Trask L, editors. Time Depth in Historical Linguistics. The McDonald Institute for Archaeological Research Press; Cambridge: 2000. pp. 143–165. [Google Scholar]
- 35.Fedorov YA. Historical ethnography of the North Caucasus. Moscow University Press; Moscow: 1983. [Google Scholar]
- 36.Fenner JN. Cross-cultural estimation of the human generation interval for use in genetics-based population divergence studies. Am J Phys Anthropol. 2005;128:415–423. doi: 10.1002/ajpa.20188. [DOI] [PubMed] [Google Scholar]
- 37.Flores C, Maca-Meyer N, Larruga JM, et al. Isolates in a corridor of migrations: a high-resolution analysis of Y-chromosome variation in Jordan. J Hum Genet. 2005;50:435–441. doi: 10.1007/s10038-005-0274-4. (6 co-authors) [DOI] [PubMed] [Google Scholar]
- 38.Forster P, Harding R, Torroni A, et al. Origin and evolution of Native American mtDNA variation: a reappraisal. Am. J. Hum. Genet. 1996;59:935–945. (4 co-authors) [PMC free article] [PubMed] [Google Scholar]
- 39.Ge J, Budowle B, Aranda XG. Mutation rates at Y chromosome short tandem repeats in Texas populations. Forensic Sci. Int. Genet. 2009;3:179–184. doi: 10.1016/j.fsigen.2009.01.007. (6 co-authors) [DOI] [PubMed] [Google Scholar]
- 40.Gell-Mann M, Peiros I, Starostin SA. Aspects of Comparative Linguistics, v.3. RSUH Publishers; Moscow: 2008. Lexicostatistics Compared with Shared Innovations: the Polynesian Case; pp. 13–44. [Google Scholar]
- 41.Gerasimova MM, Rud’ NM, Yablonskij LT. Anthropology of ancient and medieval populations of Eastern Europe. Nauka; Moscow: 1987. Anthropological data to the issue of ethnic relations in the North-East Black Sea (Bosporus kingdom) pp. 79–82. [Google Scholar]
- 42.Gigeneishvili BK. Comparative phonetics of the Dagestan languages. Tbilisi University Press; Tbilisi: 1977. [Google Scholar]
- 43.Gray RD, Atkinson QD. Language-tree divergence times support the Anatolian theory of Indo-European origins. Nature. 2003;426:435–39. doi: 10.1038/nature02029. [DOI] [PubMed] [Google Scholar]
- 44.Greenhill SJ, Atkinson QD, Meade A, et al. The shape and tempo of language evolution. Proc. Biol. Sci. 2010;277:2443–2450. doi: 10.1098/rspb.2010.0051. (4 co-authors) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gusmao L, Sánchez-Diz P, Calafell F, et al. Mutation rates at Y chromosome specific microsatellites. Hum. Mutat. 2005;26:520–528. doi: 10.1002/humu.20254. (42 co-authors) [DOI] [PubMed] [Google Scholar]
- 46.Haber M, Platt DE, Badro DA, et al. Influences of history, geography and religion on genetic structure: the Maronites in Lebanon. Eur. J. Hum. Genet. 2010 doi: 10.1038/ejhg.2010.177. (13 co-authors) (forthcoming) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.King TE, Jobling MA. Founders, Drift, and Infidelity: The Relationship between Y Chromosome Diversity and Patrilineal Surnames. Mol Biol Evol. 2009;26:1093–1102. doi: 10.1093/molbev/msp022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kitchen A, Ehret C, Assefa S, et al. Bayesian phylogenetic analysis of Semitic languages identifies an Early Bronze Age origin of Semitic in the Near East. Proc. Biol. Sci. 2009;276:2703–2710. doi: 10.1098/rspb.2009.0408. (4 co-authors) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kovalevskaya VB. North Caucasians antiquity. In: Rybakov BA, editor. The Eurasian steppes in the Middle Ages. Nauka; Moscow: 1981. pp. 83–97. (Series Archaeology of the USSR). [Google Scholar]
- 50.Kuipers AN. Caucasian. In: Sebeok T, editor. Current Trends in Linguistics. Soviet and East European Linguistics. Mouton; The Hague: 1963. pp. 315–344. [Google Scholar]
- 51.Kutuev IA, Litvinov SS, Khusainova RI, et al. The genetic structure and molecular phylogeography of Caucasus populations based on Y chromosome data. Medicinskaya Genetika. 2010;3:18–25. (5 co-authors) [Google Scholar]
- 52.Manni F, Guerard E. Barrier vs. 2.2. (computer program) Population genetics team, Museum of Mankind (Musee de l’Homme); Paris: 2004. [Google Scholar]
- 53.Masson VM, Merpert NY, Munchaev RM, et al. In: Chalcolithic of the USSR. Rybakov BA, editor. Nauka; Moscow: 1994. (Series Archaeology of the USSR). (4 co-authors) [Google Scholar]
- 54.Melyukova AI. Conclusions. In: Rybakov BA, editor. The steppes of the European part of the USSR in the Scythian-Sarmatian time. Nauka; Moscow: 1989. pp. 292–295. (Series Archaeology of the USSR). [Google Scholar]
- 55.Mudrak OA. Aspects of Comparative Linguistics. RSUH Publishers; Moscow: 2008. Kamchukchee/Eskimo Glottochronology and Some Altaic-Eskimo Etymologies Found on the Swadesh List; pp. 297–336. [Google Scholar]
- 56.Munchaev RM. Maikop culture. In: Rybakov BA, editor. The Bronze Age of the Caucasus and Central Asia. Nauka; Moscow: 1994. pp. 158–225. (Series Archaeology of the USSR). [Google Scholar]
- 57.Nasidze I, Ling EY, Quinque D, et al. Mitochondrial DNA and Y-chromosome variation in the caucasus. Ann. Hum. Genet. 2004;68:205–221. doi: 10.1046/j.1529-8817.2004.00092.x. (17 co-authors) [DOI] [PubMed] [Google Scholar]
- 58.Nasidze I, Quinque D, Dupanloup I, et al. Genetic evidence concerning the origins of South and North Ossets. Ann. Hum. Genet. 2004;68:588–599. doi: 10.1046/j.1529-8817.2004.00131.x. (7 co-authors) [DOI] [PubMed] [Google Scholar]
- 59.Nasidze I, Sarkisian T, Kerimov A, et al. Testing hypotheses of language replacement in the Caucasus: evidence from the Y-chromosome. Hum. Genet. 2003;112:255–261. doi: 10.1007/s00439-002-0874-4. (4 co-authors) [DOI] [PubMed] [Google Scholar]
- 60.Nikolaev SL, Starostin SA. A North Caucasian Etymological Dictionary. Asterisk Publishers; Moscow: 1994. [Google Scholar]
- 61.Pocheshkhova EA. Structure of migrations and gene drift in populations of Adyghes-Shapsugs. Medicinskaya Genetika. 2008;7:30–38. [Google Scholar]
- 62.Powell R, Gannon F. Purification of DNA by phenol extraction and ethanol precipitation. Oxford University Press; New York: 2002. [Google Scholar]
- 63.Rosser ZH, Zerjal T, Hurles ME, et al. Y-chromosomal diversity in Europe is clinal and influenced primarily by geography, rather than by language. Am. J. Hum. Genet. 2000;67:1526–1543. doi: 10.1086/316890. (62 co-authors) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ruhlen MA. Guide to the World’s Languages. Classification. V. 1. Stanford University Press; Stanford: 1987. [Google Scholar]
- 65.Saillard J, Forster P, Lynnerup N, et al. MtDNA variation among Greenland Eskimos: the edge of the Beringian expansion. Am. J. Hum. Genet. 2000;67:718–726. doi: 10.1086/303038. (5 co-authors) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sánchez-Diz P, Alves C, Carvalho E, et al. Population and segregation data on 17 Y-STRs: results of a GEP-ISFG collaborative study. Int. J. Legal. Med. 2008;122:529–533. doi: 10.1007/s00414-008-0265-z. (18 co-authors) [DOI] [PubMed] [Google Scholar]
- 67.Schneider S, Roessli D, Excoffier L. Arlequin vers. 2.000: a software for population genetics data analysis. Genetics and Biometry Laboratory, Department of Anthropology and Ecology. Univ. of Geneva; Geneva, Switzerland: 2000. [Google Scholar]
- 68.Semino O, Magri C, Benuzzi G, et al. Origin, diffusion, and differentiation of Y-chromosome haplogroups E and J: inferences on the neolithization of Europe and later migratory events in the Mediterranean area. Am. J. Hum. Genet. 2004;74:1023–1034. doi: 10.1086/386295. (16 co-authors) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Semino O, Passarino G, Oefner PJ, et al. The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective. Science. 2000;290:1155–1159. doi: 10.1126/science.290.5494.1155. (17 co-authors) [DOI] [PubMed] [Google Scholar]
- 70.Sengupta S, Zhivotovsky LA, King R, et al. Polarity and Temporality of High-Resolution Y-Chromosome Distributions in India Identify Both Indigenous and Exogenous Expansions and Reveal Minor Genetic Influence of Central Asian Pastoralists. Am. J. Hum. Genet. 2006;78:202–221. doi: 10.1086/499411. (15 co-authors) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shagirov AK. The etymological dictionary of Adyghe (Circassian) languages. Nauka; Moscow: 1977. [Google Scholar]
- 72.Starostin GS. A Lexicostatistical Approach Towards Reconstructing Proto-Khoisan. Mother Tongue. 2003;v. VIII:81–126. [Google Scholar]
- 73.Starostin SA. Time Depth in Historical Linguistics. The McDonald Institute for Archaeological Research Press; Cambridge: 2000. Comparative-Historical Linguistics and Lexicostatistics; pp. 223–265. [Google Scholar]
- 74.Starostin SA. Linguistic reconstruction and the ancient history of the East. Nauka; Moscow: 1989. Comparative-historical linguistics and lexicostatistics; pp. 2–39. [Google Scholar]
- 75.StatSoft, Inc STATISTICA (data analysis software system) (version 6) 2001 www.statsoft.com.
- 76.Swadesh M. Towards greater accuracy in lexicostatistic dating. International Journal of American Linguistics. 1955;21:121–137. [Google Scholar]
- 77.Talibov BB. Comparative phonetics of Lezghins languages. Nauka; Moscow: 1980. [Google Scholar]
- 78.Tofanelli S, Ferri G, Bulayeva K, et al. J1-M267 Y lineage marks climate-driven pre-historical human displacements. Eur. J. Hum. Genet. 2009;17:1520–1524. doi: 10.1038/ejhg.2009.58. (23 co-authors) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Trubetzkoy NS. Wiener Zeitschrift für die Kunde des Morgenlandes. Heft 2. Bd XXXVII. Wien; 1930. Nordkaukasische Wortgleichungen. [Google Scholar]
- 80.Underhill PA, Myres NM, Rootsi S, et al. Separating the post-Glacial coancestry of European and Asian Y chromosomes within haplogroup R1a. Eur. J. Hum. Genet. 2010;18:479–484. doi: 10.1038/ejhg.2009.194. (34 co-authors) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vogel F, Motulsky AG. Problems and Approaches. Springer-Verlag; Berlin: 1986. Human Genetics. [Google Scholar]
- 82.Wells RS, Yuldasheva N, Ruzibakiev R, et al. The Eurasian heartland: a continental perspective on Y-chromosome diversity. Proc. Natl. Acad. Sci. U S A. 2001;98:10244–10249. doi: 10.1073/pnas.171305098. (27 co-authors) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wilson IJ, Weale ME, Balding DJ. Inferences from DNA data: population histories, evolutionary processes and forensic match probabilities. J. R. Statist. Soc. 2003;166:155–201. [Google Scholar]
- 84.Womble WH. Science. 1951;114:315–322. doi: 10.1126/science.114.2961.315. [DOI] [PubMed] [Google Scholar]
- 85.Xue Y, Zerjal T, Bao W, et al. Modelling Male Prehistory in East Asia using BATWING. In: Matsumura S, Forster P, Renfrew C, editors. Simulations, genetics and prehistory: a focus on islands. McDonald Institute; Cambridge: UK: 2008. pp. 81–90. (13 co-authors) [Google Scholar]
- 86.Zalloua PA, Platt DE, El Sibai M, et al. Identifying genetic traces of historical expansions: Phoenician footprints in the Mediterranean. Am. J. Hum. Genet. 2008;83:633–642. doi: 10.1016/j.ajhg.2008.10.012. (41 co-authors) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zalloua PA, Xue Y, Khalife J, et al. Y-chromosomal diversity in Lebanon is structured by recent historical events. Am. J. Hum. Genet. 2008;82:873–882. doi: 10.1016/j.ajhg.2008.01.020. (34 co-authors) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zerjal T, Wells RS, Yuldasheva N, Ruzibakiev R, Tyler-Smith C. A genetic landscape reshaped by recent events: Y-chromosomal insights into central Asia. Am J Hum Genet. 2002;71:466–482. doi: 10.1086/342096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhivotovsky LA, Underhill PA, Cinnioğlu C, et al. The effective mutation rate at Y chromosome short tandem repeats, with application to human population-divergence time. Am. J. Hum. Genet. 2004;74:50–61. doi: 10.1086/380911. (17 co-authors) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.