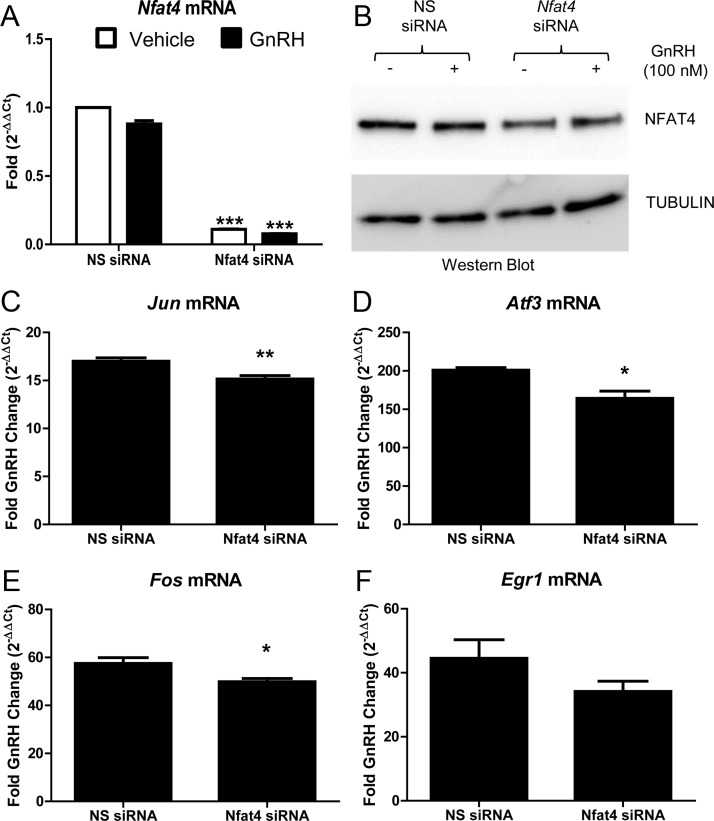
Fig. 6.
Nfat4 is required for maximal Jun, Fos, and Atf3 mRNA accumulation downstream of GnRH. LβT2 cells were transfected with a scrambled sequence NS siRNA or siRNA specific for Nfat4. Seventy-two hours after transfection, serum-starved LβT2 cells were treated with vehicle (PBS) or GnRH (100 nm) for 1 h. After treatment, RNA was isolated and reverse transcribed, and real-time PCR was performed using primers specific for Nfat4 mRNA and the IEG Jun, Fos, Atf3, and Egr1 (A and C–F). Fold changes for cells treated with GnRH and NS siRNA were compared with fold changes of cells treated with GnRH and Nfat4 siRNA using the comparative (CT) method described by Schmittgen and Livak (66). For example, the equation below was used to determine the GnRH induced fold change (2-ΔΔCt) in Jun mRNA in cultures transfected with NS siRNA: 2-ΔΔCt (Jun/NS siRNA) where ΔΔCT = [(CT Jun − CT Gapdh internal control) [GnRH treated] − (CT Jun − CT Gapdh internal control) [vehicle treated]]. 2-ΔΔCt values for each IEG mRNA in cultures transfected with either NS siRNA or Nfat siRNA are the mean ± sem from four independent experiments performed consecutively on different days. qRT-PCR samples were run in duplicate. Data were analyzed via a two-tailed unpaired t test. *, P < 0.05; **, P < 0.01. B, Whole-cell lysates from LβT2 cells transfected with siRNA were run on a SDS polyacrylamide gel and then immunoblotted for NFAT4 and tubulin, used as a loading control. These transfections were done together, with a subset collected for RNA and another for protein as described in Materials and Methods.