Abstract
The activation of phospholipase D (PLD) produces phosphatidic acid (PA), whereas plant sphingosine kinase (SPHK) phosphorylates long-chain bases to generate long-chain base-1-phosphates such as phytosphingosine-1-phosphate (phyto-S1P). PA and phyto-S1P have been identified as lipid messengers. Recent studies have shown that PA interacts directly with SPHKs in Arabidopsis, and that the interaction promotes SPHK activity. However, SPHK and phyto-S1P act upstream of PLDα1 and PA in the stomatal response to abscisic acid (ABA). These findings indicate that SPHK/phyto-S1P and PLD/PA are co-dependent in the amplification of lipid messengers, and that crosstalk between the sphingolipid- and phospholipid-mediated signaling pathways may play important roles in plant stress signaling.
Keywords: phospholipase D, phosphatidic acid, sphingosine kinase, phytosphingosine, lipid signaling, abscisic acid
Introduction
Different classes of lipids have been implicated as lipid messengers in plant growth, development, and stress responses, and recent results have begun to unveil complex interactions among different lipid signaling pathways (Peters et al., 2010; Guo et al., 2011). Under a given stress, more than one lipid mediators are often produced, with some being antagonistic and others having similar functions. Both phosphatidic acid (PA) and long-chain base-1-phosphate (LCBP) promote abscisic acid (ABA)-mediated stomatal closure and decrease reactive oxygen species (ROS)-induced cell death (Jacob et al., 1999; Zhang et al., 2003; Coursol et al., 2005; Shi et al., 2007). ABA and ROS are pivotal signals impacting various aspects of plant growth and stress responses. This raises intriguing questions of how these two lipid signaling processes interact to mediate plant stress responses. Recent results indicate a crosstalk between phospholipase D (PLD) and sphingosine kinase (SPHK) during the production of lipid messengers. These interactions of phospholipid- and sphingolipid-mediated signaling pathways may play important roles in plant response to various stresses.
Different PLDs Involved in Diverse Stress Responses
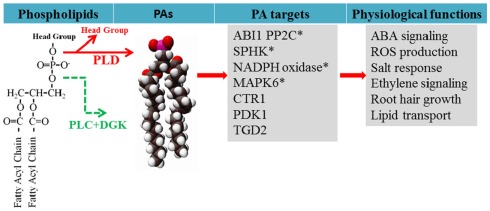
Phospholipase D hydrolyzes phospholipids to produce PA and a free head group (Figure 1). This enzyme was first discovered in plants and has since been found to occur also in bacteria, fungi, and animals (Wang et al., 1994; Qin et al., 1997; Wang, 2001). The Arabidopsis genome has 12 genes encoding PLDs, which are grouped into six classes, PLDα(1–3), β(1, 2), γ(1–3), δ, ε, and ζ(1, 2) based on the gene sequences, protein domain structures, and enzymatic biochemical properties (Wang et al., 2006). PLDα, β, γ, δ, and ε contain a Ca2+/phospholipids-binding C2 domain whereas PLDζ1 and ζ2 contain the pleckstrin homology (PH) and phox homology (PX) domain (Wang et al., 2006). All the PLDs have two conserved HxKxxxD (HKD) motifs that are involved in catalytic activities (Wang et al., 2006; Li et al., 2009). Some of the C2-containing PLDs contain a polyphosphoinositide-binding region (PBR1) located between two HKD domains, which binds phosphatidylinositol 4,5-bisphosphate (PIP2; Zheng et al., 2002).
Figure 1.
Generation of PA from phospholipids and PA target proteins functionally characterized in plants. PA is generated via two pathways during stress responses: PLD hydrolyzes phospholipid to generate PA, and PLC hydrolyzes phospholipid to generate DAG which can be phosphorylated by DAG kinase (DGK). PA has been found to interact with target proteins to regulate cellular functions. Examples of PA regulation of target proteins are discussed in the text and the references are cited in the text. *Indicates that PA is generated from PLD while others are not determined.
These sequence differences provide a structural basis for distinctively different biochemical properties for different PLDs. All the C2-containing PLDs require Ca2+ for activity, but PX and PH-containing PLDζs do not (Wang et al., 2006). In addition, the differences in the C2 sequences can explain in part, the different Ca2+ concentration requirements. PLDα1 is most active when assayed at millimolar [Ca2+] whereas PLDβ1 and PLDγ1 require micromolar concentrations of Ca2+ for optimal activity and also require PIP2 as a co-factor (Qin et al., 1997; Zheng et al., 2002; Pappan et al., 2004). PLDδ and PLDε both are active within a broad range of Ca2+ concentrations (μM–mM; Hong et al., 2008, 2009). PLDδ requires oleate and PIP2 for its activity, but PLDε is active under the reaction conditions of PLDα1, β1, γ1, and δ (Wang and Wang, 2001; Qin et al., 2002; Hong et al., 2008). Arabidopsis PLDs also selectively hydrolyze common membrane phospholipids such as PC, PE, and PG (Li et al., 2009). The varied co-factor requirements and substrate preferences for different PLDs indicate that specific PLDs are activated differently in the cell, and may have unique cellular and physiological functions (Li et al., 2009).
Different PLDs are involved in various physiological processes, displaying unique and overlapping functions (Figure 1; Li et al., 2009). PLDα1-deficient plants have an altered plant response to several stresses, including water loss (Sang et al., 2001a), ROS production (Sang et al., 2001b; Zhang et al., 2009), and salt tolerance (Bargmann et al., 2009; Yu et al., 2010). PLDδ is involved in freezing tolerance (Li et al., 2004), dehydration (Katagiri et al., 2001), salt tolerance (Bargmann et al., 2009), H2O2-induced programmed cell death (PCD; Zhang et al., 2003), microtubule organization, and cytoskeletal rearrangement (Gardiner et al., 2001, 2003). PLDα3 is also involved in salt tolerance (Hong et al., 2008) whereas PLDε enhances Arabidopsis nitrogen signaling and growth (Hong et al., 2009). PLDζ1 and ζ2 are involved in lipid remodeling and root growth in plant responses to phosphate deprivation (Cruz-Ramirez et al., 2006; Li et al., 2006a,b). PLDζ1 is implicated in root-hair patterning (Ohashi et al., 2003), and PLDζ2 participates in vesicle trafficking to regulate auxin response (Li and Xue, 2007).
PA as a Pivotal Class of Lipid Messengers
One mechanism by which PLDs affect plant stress responses is to produce PA, which has been identified as a class of lipid messengers in plants and animals (Figure 1). PA constitutes less than 1% of total phospholipids in most plant tissues, but the cellular level of PA changes dynamically in plants under abiotic and biotic stresses (Wang et al., 2006). The amount of PA in Arabidopsis leaves increased more than 60% within 10 min of application of ABA (Zhang et al., 2004). Other stresses, including wounding, freezing, various osmotic stresses, oxidative stress, and drought, induce accumulation of PA (Li et al., 2009). Manipulations of various PLDs in Arabidopsis have shed light on the regulatory functions of PA. Characterization of knockouts, knockdown, and overexpression lines of PLDs, has shown that PA produced from different PLDs has unique roles in plant response to different stresses, including water deficits, high salinity, freezing, phosphate deprivation, nitrogen availability, and plant-pathogen interactions (Sang et al., 2001b; Zhang et al., 2003; Hong et al., 2008, 2009; Bargmann et al., 2009; Peters et al., 2010).
One mode of PA action is its direct interaction with target proteins (Figure 1). In yeast and animal cells, PA binds to transcriptional factors, protein kinases, lipid kinases, protein phosphatases, and proteins involved in vesicular trafficking and cytoskeletal rearrangement (Wang et al., 2006; Gomez-Cambronero, 2010). In plants, PA has been found to interact with ABI1 PP2C phosphatase (Zhang et al., 2004), phosphoinositide-dependent protein kinase1 (Anthony et al., 2004), phosphoenolpyruvate carboxylase (Testerink et al., 2004), CTR1 protein kinase (Testerink et al., 2007), the actin capping protein AtCP (Huang et al., 2006), lipid transport protein TGD2 (Lu and Benning, 2009), NADPH oxidase (Zhang et al., 2009), mitogen-activated protein kinase 6 (Yu et al., 2010), and SPHK (Guo et al., 2011; Figure 1). Several potential PA-interacting proteins were identified by PA-affinity chromatography followed by mass spectrometric analyses in plants (Testerink et al., 2004). PA-protein interaction may modulate the function of a protein in two ways, tethering it to the membrane to change their localization, and/or increasing or decreasing the enzyme catalytic activity. For example, PLDα1-derived PA interacts with ABI1 and tethers ABI1 to the plasma membrane (Zhang et al., 2004). PA binds to Arabidopsis NADPH oxidase and SPHK to promote their activity (Zhang et al., 2009; Guo et al., 2011).
In addition to PLD, signaling PA can be produced by the diacylglycerol (DAG) kinase phosphorylation of DAG, which is often produced by the activation of phospholipase C (PLC; Figure 1). Two distinctively different PLC families have been described in plants, the phosphoinositol 4,5-bisphosphate-hydrolyzing PI-PLC (Munnik, 2001) and the non-specific PLC (NPC) that hydrolyze common membrane phospholipids such as PC and PE (Peters et al., 2010). It should be noted that DAG itself can serve as a lipid mediator; DAG promotes stomatal opening (Lee and Assmann, 1991; Peters et al., 2010), whereas PA promotes stomatal closure (Jacob et al., 1999; Zhang et al., 2004; Mishra et al., 2006).
SPHKs in Plants
Sphingosine kinase is a member of the DAG kinase family (Strub et al., 2010), and phosphorylates long-chain bases (LCBs) to LCBPs, such as sphingosine-1-phopshate (S1P) and phyto-S1P (Figure 2A). SPHK activity and function have been well characterized in animals and yeast (Worrall et al., 2003). In mammals, two SPHKs and their product S1P have important roles in regulation of many cellular processes including cell growth, suppression of apoptosis, and pathophysiology of various diseases (Strub et al., 2010). While sphingosine (d18:1Δ4) is the predominant LCB in animal cells, it is only detected as a minor LCB in some plants or absent in other plants, such as Arabidopsis (Lynch et al., 2009; Michaelson et al., 2009). A recent survey of 21 species from different phylogenetic groups has found that d18:1Δ4 is present in non-seed land plants and monocots (wheat, barley, maize, and ryegrass), but it is absent in Arabidopsis and soybean (Islam et al., 2012). Instead, 4-hydroxy-sphingenine (t18:0, commonly known as phytosphingosine), 4-hydroxy-8-sphingenine (t18:1Δ8), and 8-sphingenine (d18:1Δ8) are predominant LCBs in plants (Lynch et al., 2009). Plant extracts and purified SPHKs phosphorylate various LCBs to generate LCBPs (Coursol et al., 2005; Guo et al., 2011).
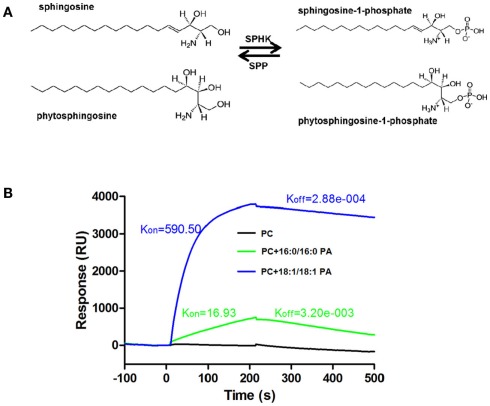
Figure 2.
Phosphorylation of sphingosine and phytosphingosine by SPHK and the interaction SPHK and PA. (A) SPHK catalyzes the formation of S1P or phyto-S1P from sphingosine or phytosphingosine. S1P or phyto-S1P can be degraded by S1P phosphatase (SPP) or S1P lyase (not shown). (B) Surface plasmon resonance (SPR) analysis of interaction of PA with SPHK1. Liposomes containing PC only or PC plus 16:0/16:0 or 18:1/18:1 PA were used to analyze the interaction. Liposome containing of PC did not bind to SPHK1. Liposomes containing both PC and PA (16:0/16:0 or 18:1/18:1) bound to SPHK1. (B) is based on data from Guo et al., 2011.
The Arabidopsis genome contains five genes with sequence similarities to mammalian SPHKs. At5g23450 encodes a LCB kinase AtLCBK1 (Nishiura et al., 2000; Imai and Nishiura, 2005) whereas At5g51290 is regarded as a ceramide kinase (Liang et al., 2003). At2g46090 did not have sphingosine-phosphorylating activity (Worrall et al., 2008). At4g21540 was originally annotated as one SPHK, and this sequence consists of two repeats that are most similar to mammalian SPHKs. A cDNA from the second repeat was reported to encode an active SPHK, designated SPHK1 (Worrall et al., 2008). A recent study has established that the At4g21540 locus is actually comprised of two separate SPHK genes, SPHK1 and SPHK2. The conclusion is supported by molecular cloning, sequence analysis, and the distinguishable patterns of expression of SPHK1 and SPHK2 in Arabidopsis tissues (Guo et al., 2011). The stop codon of SPHK2 is 788 bp upstream of the start codon of SPHK1. Both SPHK1 and SPHK2 were localized on tonoplasts (Worrall et al., 2008; Guo et al., 2011). SPHK1, SPHK2, and AtLCBK1 utilize various LCBs as substrates with different preference. Among the substrates tested, AtLCBK1 prefers d-erythro-dihydrosphingosine to sphingosine and phytosphingosine, whereas SPHK1 and SPHK2 are most active on sphingosine. AtLCBK1 cannot phosphorylate D-threo-dihydrosphingosine (Imai and Nishiura, 2005) but both SPHK1 and 2 can even though SPHK2 has much a lower activity than SPHK1 (Guo et al., 2011). Because of the low occurrence of sphingosine in plant tissues and the broad substrate specificity of SPHKs, it was suggested that plant SPHKs should be called LCB kinase (LCBK) in plants (Lynch et al., 2009). This change will require renaming some of the genes in the family. SPHK1 and SPHK2 are used here for consistency with published nomenclature on these enzymes (Worrall et al., 2008; Guo et al., 2011, 2012).
LCBs as Lipid Mediators
Like glycerophospholipids, sphingolipids serve not only as a main component of cell membranes, but also important signaling molecules (Lynch et al., 2009; Pata et al., 2010). S1P is produced in animal cells by two SPHKs and is degraded either by S1P lyase or S1P phosphatases (Figure 2A). S1P regulates a variety of developmental and disease processes in animals (Strub et al., 2010). Many lines of evidence indicate that S1P is an intracellular messenger acting directly on intracellular target proteins (Maceyka et al., 2012). In addition, S1P is exported out of cells to mediate signaling pathways through five specific G protein-coupled receptors (S1RP1–S1RP5) on the plasma membrane (Maceyka et al., 2012).
Sphingolipids are emerging as important mediators in plants and accumulating evidence indicates that sphingolipid metabolites, including LCBs, LCBPs, and ceramides, are involved in various signaling pathways in plants (Lynch et al., 2009; Pata et al., 2010). Characterization of Arabidopsis deficient in sphingolipid metabolism genes facilitates the understanding of signaling and physiological functions of sphingolipid in plants. The key roles of sphingolipids in PCD have been extensively investigated (Berkey et al., 2012). For example, characterization of ceramide kinase mutant (acd5) shows that ceramide induces plant PCD whereas phosphorylated ceramide partially attenuates PCD (Liang et al., 2003). Recent studies suggest that both LCB and LCBP are involved in PCD (Shi et al., 2007; Alden et al., 2011). Mutation of a LCB1 subunit of serine palmitoyltransferase blocks accumulation of LCBs in Arabidopsis and indicates that LCBs are involved in initiating PCD through induction of ROS production in Arabidopsis (Shi et al., 2007; Wang et al., 2008). LCBPs have been shown to decrease ROS-induced PCD whereas unphosphorylated LCBs promote ROS-mediated cell death (Shi et al., 2007). LCB-induced ROS production is also found to depend on NADPH oxidase Respiratory Burst Oxidase Homolog D (Peer et al., 2011). Recently, a study indicates that another subunit of serine palmitoyltransferase, LCB2a, is required for PCD, and MPK6 mediates downstream signal in LCB-induced PCD (Saucedo-Garcia et al., 2011). These results suggest that the balance between unphosphorylated and phosphorylated form of sphingolipids may function as a rheostat in regulation of PCD.
SPHK/Phyto-S1P and PLD/PA Both Involved in the ABA Signaling Pathway
One of the functions that have been studied for SPHK and phyto-S1P is their roles in mediating ABA-promoted stomatal closure. ABA treatments increased SPHK activity in Arabidopsis and drought stress induced the production of LCBPs in Commelina communis (Ng et al., 2001; Coursol et al., 2003). Application of S1P induces stomatal closure and inhibits stomatal opening (Ng et al., 2001). Knockout of either SPHK1 or SPHK2 decreased the sensitivity to ABA in Arabidopsis, whereas overexpression of SPHK1 or SPHK2 increased ABA sensitivity (Worrall et al., 2008; Guo et al., 2011). The involvement of LCBP in the ABA signaling in guard cells is further supported by analysis of the LCBP phosphatase AtSPP1 mutant spp1 (Figure 2A). AtSPP1 is suggested to be involved in regulation of LCBP level during ABA response. The spp1 plants displayed increased sensitivity to ABA in stomatal closure due to a defect in LCBP degradation in the mutant (Nakagawa et al., 2011). Thus, LCBP levels regulated by SPHKs and AtSPP1 may play an important role in the ABA signaling pathway.
Likewise, a number of studies have shown that PLD and PA play important roles in signaling ABA-mediated stomatal closure (Jacob et al., 1999; Zhang et al., 2004). PLD and PA promote open stomata to close and meanwhile prevent the closed stomata from opening (Jacob et al., 1999; Zhang et al., 2004). In Arabidopsis, PLDα1-deficient plants displayed insensitivity to ABA, whereas overexpression (OE) of PLDα1 resulted in increased sensitivity to ABA (Sang et al., 2001a). PLDα1 regulates ABA signaling pathways through different interactions (Figure 3). PA binds to ABI1 phosphatase 2C, and this interaction inhibits the negative function of ABI1 in ABA response and mediates ABA-promoted stomatal closure (Zhang et al., 2004; Mishra et al., 2006). On the other hand, PLDα1 interacts with Gα to mediate the ABA inhibition of stomatal opening (Zhao and Wang, 2004; Mishra et al., 2006). In addition, PLDα1-derived PA binds to and increases NADPH oxidase activity to promote the production of ROS in ABA-mediated stomatal closure (Figure 1; Zhang et al., 2009).
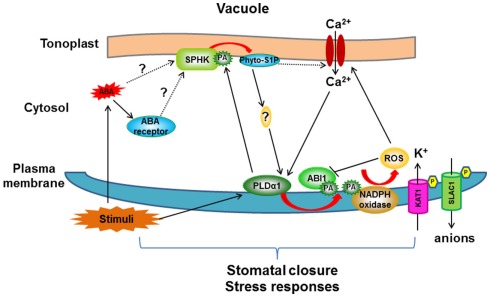
Figure 3.
Proposed model for crosstalk between PLDα1/PA and SPHK/phyto-S1P in ABA-mediated stomatal closure signaling pathway. ABA may be perceived by the receptor (PYR/PYL/RCAR) in the cytosol, leading to activation of SPHK to produce phyto-S1P which initiates a cascade to activate PLDα1. PLDα1 hydrolyzes phospholipids to increase PA level in membrane (plasma membrane and tonoplast). PLDα1-deprived PA promotes the ABA effect through three targets: (i) PA binds to ABI1 and tethers ABI1 to the membrane to inhibit its negative effect; (ii) PA stimulates plasma membrane-localized NADPH oxidase to form secondary messenger: ROS; (iii) Increased PA in tonoplast interacts with SPHK and promotes its activity to form a positive loop. PLDα1/PA- and SPHK/phyto-S1P-mediated signaling pathway activates ion channel activity, leading to ion flux in guard cell and finally stomatal closure. Note that this model summarizes the crosstalk between PLDα1/PA and SPHK/phyto-S1P and their roles in ABA-mediated stomatal closure, not all ABA signaling components are included in this model. Arrow indicates positive regulation, bar indicates repression. Red arrow represents reactions which produce secondary signaling molecules.
PA Interaction with SPHK to Promote LCBP Production
The findings that both PLD/PA and SPHK/phyto-S1P are involved in stomatal closure raise an intriguing question of whether the two lipid signaling processes interact to mediate plant responses to ABA and stress. A recent study investigated the direct interaction of PA with two Arabidopsis SPHKs (Guo et al., 2011). PA binds to both Arabidopsis SPHKs and the interaction stimulates SPHK activity. The interaction was demonstrated by different approaches, including lipid-filter binding, liposome binding, surface plasmon resonance (SPR), and validated using PA-SPHK co-precipitation from protoplasts (Figure 2B; Guo et al., 2011, 2012). PA has various molecular species which differ in acyl chain length and degree of saturation. PAs with 18:1/18:1, 16:0/18:1, and 16:0/18:2 acyl chains bind strongly to both SPHKs, whereas 16:0/16:0, 8:0/8:0, 18:0/18:0, and 18:2/18:2 PAs bind poorly to SPHKs (Guo et al., 2011).
The identification of SPHKs as molecular targets of PA indicates that PA may mediate the ABA activation of SPHK in plants. Indeed, in response to ABA, the LCBP level is lower in pldα1. In addition, the application of PA increased the LCBP production in protoplasts (Guo et al., 2012). These results are consistent with the hypothesis that SPHK activation by ABA is mediated by PA. On the other hand, in response to ABA, the PA production in sphk1-1 and sphk2-1 was significantly lower than WT while overexpression of SPHK increased PA production, suggesting that PLDα1 activation depends on SPHK (Guo et al., 2012). Taken together, these results indicate a co-dependence of PLD/PA and SPHK/phyto-S1P in the production of PA and phyto-S1P lipid messengers (Figure 3).
SPHK/LCBP Acting Upstream of PLD/PA
To delineate the signaling steps of PLDα1 and SPHKs in the ABA signaling, PA and phyto-S1P were supplemented to the epidermal peels of PLDα1 or SPHK-deficient plants. PA promoted stomatal closure in PLDα1-KO or SPHK-KO leaves, whereas phyto-S1P promoted stomatal closure in SPHK-KO but not in PLDα1-KO mutant. Furthermore, the addition of 1-butanol, which suppresses PA production by PLD, attenuated the effect of phyto-S1P-induced stomatal closure (Guo et al., 2012). These results suggest that phyto-S1P-mediated stomatal closure requires PLDα1, and that SPHK/phyto-S1P acts upstream of PLDα1.
These enzymatic, genetic, physiological, and lipid analyses indicate a positive interplay between the two lipid signaling processes, SPHK/phyto-S1P and PLD/PA, in plant response to stresses (Figure 3). ABA is produced under various stresses, such as drought and high salinity. ABA activates SPHKs to generate phyto-S1P which promotes the activation of PLDα1, possibly through increasing the cytoplasmic Ca2+ concentration (Figure 3). PA produced by activated PLDα1 binds to SPHK and promotes SPHK activity, forming a positive feedback loop in response to ABA. The resulting increase in PA regulates downstream proteins including ABI1 and NADPH oxidase in ABA-mediated stomatal closure (Zhang et al., 2004, 2009; Mishra et al., 2006; Figure 3).
The interplay between PLDα1 and SPHK provides insights to a mechanism by which stress signaling events are communicated between the plasma and vacuolar membranes (Figure 3). The subcellular localization of membrane-based lipid signaling is expected to play an important role in regulation of enzyme activation, generation of lipid messengers, and mediation of downstream events (Li et al., 2009). It is not well understood how signaling events between different subcellular compartments are coordinated. In animals, acidic phospholipids including PA have been shown to stimulate SPHK activity (Olivera et al., 1996). PA has been implicated in promoting the intracellular translocation of cytosolic murine SPHK1 to membrane regions that are enriched in PA (Delon et al., 2004). By comparison, SPHKs in Arabidopsis are already associated with tonoplasts, and surface dilution kinetics analysis indicates that PA stimulates SPHK activity by promoting the substrate binding to the catalytic site of SPHK (Guo et al., 2011). At present, the source and level of free LCBs in the tonoplast are unknown. LCBs are synthesized in the ER and the level of free LCBs is very low in Arabidopsis. It might be possible that LCBs are released from the catabolism of complex sphingolipids by ceramidase activity. In addition, the level of free LCBs may increase via de novo biosynthesis in response to stimuli. For example, infection of Pseudomonas syringae triggered de novo synthesis of phytosphingosine from sphinganine and phytosphingosine constitutes about 5–8% of total LCBs in Arabidopsis leaves (Lynch et al., 2009; Maceyka et al., 2012).
PLDα1 is present in both the soluble and membrane fractions and it translocates from the cytosol to membranes in response to stress (Ryu and Wang, 1998; Fan et al., 1999). In response to ABA, SPHK is activated to produce phyto-S1P (possibly along with other LCBPs) on the vacuolar membrane. Phyto-S1P does not activate PLDα1 directly in vitro (Guo et al., 2012). It was shown that S1P caused an increase in Ca2+ in response to ABA (Ng et al., 2001), and thus phyto-S1P may increase cytoplasmic Ca2+ to promote PLDα1 translocation to the plasma membranes and tonoplasts. Ca2+ is a key factor required for PLDα1 activity (Qin et al., 1997). Ca2+ promotes PLD translocation and its binding to the C2 domain increases the protein association with membrane lipids such as PC. This membrane association activates PLD to generate PA that binds to SPHK to promote its activity, thus forming a positive feedback loop.
Perspectives
The progress in understanding the crosstalk of signaling events provides a functional arrangement of SPHK/phyto-S1P and PLD/PA in transducing ABA signals in guard cells. Meanwhile, the connections between the two lipid signaling processes raise many questions that warrant further investigations. ABA signaling involves multiple pathways and many regulatory elements. A core pathway of ABA signaling has been established: ABA binds to the receptor PYR/PYL/RCARs, leading to inhibition of negative regulator type 2C protein phosphatases such as ABI1, resulting in SNF1-related kinase 2 (SnRK2) activation involved in mediating downstream signaling (Klingler et al., 2010; Umezawa et al., 2010). How would the SPHK/phyto-S1P- and PLD/PA-mediated processes interact with the PYR/PYL/RCAR-ABI1-SnRK2 components? PA has been shown to bind to and inhibit ABI1 (Zhang et al., 2004). Would the PYR/PYL/RCAR-ABI1-SnRK2 components be involved in the activation of SPHK and/or PLD? ABA is a key stress hormone involved in plant response to various stresses. Is the crosstalk between PLDα1/PA and SPHK/phyto-S1P involved in other regulatory pathways in plant response to other stresses? Both PA and LCBP have been implicated in decreasing ROS-induced PCD, and different PLDs have been shown to promote ROS production and response (Zhang et al., 2003, 2009). PLDs are activated rapidly under various stress conditions. Would the activation of the plasma membrane-associated PLDs act upstream of SPHKs under different stresses? In addition, multiple LCB kinases, including AtLCBK1, SPHK1, and SPHK2, exist in Arabidopsis, and double and triple mutants deficient in two or more of these kinases can be made to help determine the role of these enzymes in the production and function of LCBPs in plant growth, development and response to stresses. Further study of the signaling events will lead to a better understanding of how plants adapt to stresses and changing environments.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Work from Xuemin Wang lab was supported by grants from the National Science Foundation (IOS-0818740; MCB-0922879) and the US Department of Energy (DE-SC0001295).
References
- Alden K. P., Dhondt-Cordelier S., Mcdonald K. L., Reape T. J., Ng C. K., Mccabe P. F., Leaver C. J. (2011). Sphingolipid long chain base phosphates can regulate apoptotic-like programmed cell death in plants. Biochem. Biophys. Res. Commun. 410, 574–580 10.1016/j.bbrc.2011.06.028 [DOI] [PubMed] [Google Scholar]
- Anthony R. G., Henriques R., Helfer A., Meszaros T., Rios G., Testerink C., Munnik T., Deak M., Koncz C., Bogre L. (2004). A protein kinase target of a PDK1 signalling pathway is involved in root hair growth in Arabidopsis. EMBO J. 23, 572–581 10.1038/sj.emboj.7600068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann B. O., Laxalt A. M., Ter Riet B., Van Schooten B., Merquiol E., Testerink C., Haring M. A., Bartels D., Munnik T. (2009). Multiple PLDs required for high salinity and water deficit tolerance in plants. Plant Cell Physiol. 50, 78–89 10.1093/pcp/pcn173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkey R., Bendigeri D., Xiao S. (2012). Sphingolipids and plant defense/disease: the “death” connection and beyond. Front. Plant Physiol. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coursol S., Fan L. M., Le Stunff H., Spiegel S., Gilroy S., Assmann S. M. (2003). Sphingolipid signalling in Arabidopsis guard cells involves heterotrimeric G proteins. Nature 423, 651–654 10.1038/nature01643 [DOI] [PubMed] [Google Scholar]
- Coursol S., Le Stunff H., Lynch D. V., Gilroy S., Assmann S. M., Spiegel S. (2005). Arabidopsis sphingosine kinase and the effects of phytosphingosine-1-phosphate on stomatal aperture. Plant Physiol. 137, 724–737 10.1104/pp.104.055806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Ramirez A., Oropeza-Aburto A., Razo-Hernandez F., Ramirez-Chavez E., Herrera-Estrella L. (2006). Phospholipase Dζ2 plays an important role in extraplastidic galactolipid biosynthesis and phosphate recycling in Arabidopsis roots. Proc. Natl. Acad. Sci. U.S.A. 103, 6765–6770 10.1073/pnas.0600863103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delon C., Manifava M., Wood E., Thompson D., Krugmann S., Pyne S., Ktistakis N. T. (2004). Sphingosine kinase 1 is an intracellular effector of phosphatidic acid. J. Biol. Chem. 279, 44763–44774 10.1074/jbc.M405771200 [DOI] [PubMed] [Google Scholar]
- Fan L., Zheng S., Cui D., Wang X. (1999). Subcellular distribution and tissue expression of phospholipase Dα, Dβ, and Dγ in Arabidopsis. Plant Physiol. 119, 1371–1378 10.1104/pp.119.4.1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner J., Collings D. A., Harper J. D., Marc J. (2003). The effects of the phospholipase D-antagonist 1-butanol on seedling development and microtubule organisation in Arabidopsis. Plant Cell Physiol. 44, 687–696 10.1093/pcp/pcg095 [DOI] [PubMed] [Google Scholar]
- Gardiner J. C., Harper J. D., Weerakoon N. D., Collings D. A., Ritchie S., Gilroy S., Cyr R. J., Marc J. (2001). A 90-kD phospholipase D from tobacco binds to microtubules and the plasma membrane. Plant Cell 13, 2143–2158 10.1105/tpc.13.9.2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Cambronero J. (2010). New concepts in phospholipase D signaling in inflammation and cancer. ScientificWorldJournal 10, 1356–1369 10.1100/tsw.2010.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Mishra G., Markham J. E., Li M., Tawfall A., Welti R., Wang X. (2012). Connections between sphingosine kinase and phospholipase D in the abscisic acid signaling pathway in Arabidopsis. J. Biol. Chem. 10.1074/jbc.M111.274274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Mishra G., Taylor K., Wang X. (2011). Phosphatidic acid binds and stimulates Arabidopsis sphingosine kinases. J. Biol. Chem. 286, 13336–13345 10.1074/jbc.M111.258830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y., Devaiah S. P., Bahn S. C., Thamasandra B. N., Li M., Welti R., Wang X. (2009). Phospholipase Dε and phosphatidic acid enhance Arabidopsis nitrogen signaling and growth. Plant J. 58, 376–387 10.1111/j.1365-313X.2009.03788.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y., Pan X., Welti R., Wang X. (2008). Phospholipase D(3 is involved in the hyperosmotic response in Arabidopsis. Plant Cell 20, 803–816 10.1105/tpc.107.056390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Gao L., Blanchoin L., Staiger C. J. (2006). Heterodimeric capping protein from Arabidopsis is regulated by phosphatidic acid. Mol. Biol. Cell 17, 1946–1958 10.1091/mbc.E05-12-1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai H., Nishiura H. (2005). Phosphorylation of sphingoid long-chain bases in Arabidopsis: functional characterization and expression of the first sphingoid long-chain base kinase gene in plants. Plant Cell Physiol. 46, 375–380 10.1093/pcp/pci023 [DOI] [PubMed] [Google Scholar]
- Islam M. N., Jacquemot M. P., Coursol S., Ng C. K. (2012). Sphingosine in plants – more riddles from the Sphinx? New Phytol. 193, 51–57 10.1111/j.1469-8137.2011.03963.x [DOI] [PubMed] [Google Scholar]
- Jacob T., Ritchie S., Assmann S. M., Gilroy S. (1999). Abscisic acid signal transduction in guard cells is mediated by phospholipase D activity. Proc. Natl. Acad. Sci. U.S.A. 96, 12192–12197 10.1073/pnas.96.5.1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri T., Takahashi S., Shinozaki K. (2001). Involvement of a novel Arabidopsis phospholipase D, AtPLDδ, in dehydration-inducible accumulation of phosphatidic acid in stress signalling. Plant J. 26, 595–605 10.1046/j.1365-313x.2001.01060.x [DOI] [PubMed] [Google Scholar]
- Klingler J. P., Batelli G., Zhu J. K. (2010). ABA receptors: the START of a new paradigm in phytohormone signalling. J. Exp. Bot. 61, 3199–3210 10.1093/jxb/erq151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Assmann S. M. (1991). Diacylglycerols induce both ion pumping in patch-clamped guard-cell protoplasts and opening of intact stomata. Proc. Natl. Acad. Sci. U.S.A. 88, 2127–2131 10.1073/pnas.88.14.6181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Xue H. W. (2007). Arabidopsis PLDζ2 regulates vesicle trafficking and is required for auxin response. Plant Cell 19, 281–295 10.1105/tpc.106.049965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Hong Y., Wang X. (2009). Phospholipase D- and phosphatidic acid-mediated signaling in plants. Biochim. Biophys. Acta 1791, 927–935 [DOI] [PubMed] [Google Scholar]
- Li M., Qin C., Welti R., Wang X. (2006a). Double knockouts of phospholipases Dζ1 and Dζ2 in Arabidopsis affect root elongation during phosphate-limited growth but do not affect root hair patterning. Plant Physiol. 140, 761–770 10.1104/pp.105.070987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Welti R., Wang X. (2006b). Quantitative profiling of Arabidopsis polar glycerolipids in response to phosphorus starvation. Roles of phospholipases Dζ1 and Dζ2 in phosphatidylcholine hydrolysis and digalactosyldiacylglycerol accumulation in phosphorus-starved plants. Plant Physiol. 142, 750–761 10.1104/pp.106.085647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Li M., Zhang W., Welti R., Wang X. (2004). The plasma membrane-bound phospholipase Dδ enhances freezing tolerance in Arabidopsis thaliana. Nat. Biotechnol. 22, 427–433 10.1038/nbt949 [DOI] [PubMed] [Google Scholar]
- Liang H., Yao N., Song J. T., Luo S., Lu H., Greenberg J. T. (2003). Ceramides modulate programmed cell death in plants. Genes Dev. 17, 2636–2641 10.1101/gad.1140503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B., Benning C. (2009). A 25-amino acid sequence of the Arabidopsis TGD2 protein is sufficient for specific binding of phosphatidic acid. J. Biol. Chem. 284, 17420–17427 10.1074/jbc.M109.008821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch D. V., Chen M., Cahoon E. B. (2009). Lipid signaling in Arabidopsis: no sphingosine? No problem! Trends Plant Sci 14, 463–466 10.1016/j.tplants.2009.06.008 [DOI] [PubMed] [Google Scholar]
- Maceyka M., Harikumar K. B., Milstien S., Spiegel S. (2012). Sphingosine-1-phosphate signaling and its role in disease. Trends Cell Biol. 22, 50–60 10.1016/j.tcb.2011.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson L. V., Zauner S., Markham J. E., Haslam R. P., Desikan R., Mugford S., Albrecht S., Warnecke D., Sperling P., Heinz E., Napier J. A. (2009). Functional characterization of a higher plant sphingolipid Δ4-desaturase: defining the role of sphingosine and sphingosine-1-phosphate in Arabidopsis. Plant Physiol. 149, 487–498 10.1104/pp.108.129411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra G., Zhang W., Deng F., Zhao J., Wang X. (2006). A bifurcating pathway directs abscisic acid effects on stomatal closure and opening in Arabidopsis. Science 312, 264–266 10.1126/science.1123769 [DOI] [PubMed] [Google Scholar]
- Munnik T. (2001). Phosphatidic acid: an emerging plant lipid second messenger. Trends Plant Sci. 6, 227–233 10.1016/S1360-1385(01)01918-5 [DOI] [PubMed] [Google Scholar]
- Nakagawa N., Kato M., Takahashi Y., Shimazaki K. I., Tamura K., Tokuji Y., Kihara A., Imai H. (2011). Degradation of long-chain base 1-phosphate (LCBP) in Arabidopsis: functional characterization of LCBP phosphatase involved in the dehydration stress response. J. Plant Res. 10.1007/s10265-011-0451-9 [DOI] [PubMed] [Google Scholar]
- Ng C. K., Carr K., Mcainsh M. R., Powell B., Hetherington A. M. (2001). Drought-induced guard cell signal transduction involves sphingosine-1-phosphate. Nature 410, 596–599 10.1038/35069092 [DOI] [PubMed] [Google Scholar]
- Nishiura H., Tamura K., Morimoto Y., Imai H. (2000). Characterization of sphingolipid long-chain base kinase in Arabidopsis thaliana. Biochem. Soc. Trans. 28, 747–748 10.1042/BST0280747 [DOI] [PubMed] [Google Scholar]
- Ohashi Y., Oka A., Rodrigues-Pousada R., Possenti M., Ruberti I., Morelli G., Aoyama T. (2003). Modulation of phospholipid signaling by GLABRA2 in root-hair pattern formation. Science 300, 1427–1430 10.1126/science.1083695 [DOI] [PubMed] [Google Scholar]
- Olivera A., Rosenthal J., Spiegel S. (1996). Effect of acidic phospholipids on sphingosine kinase. J. Cell. Biochem. 60, 529–537 [DOI] [PubMed] [Google Scholar]
- Pappan K., Zheng L., Krishnamoorthi R., Wang X. (2004). Evidence for and characterization of Ca2+ binding to the catalytic region of Arabidopsis thaliana phospholipase Dβ. J. Biol. Chem. 279, 47833–47839 10.1074/jbc.M402789200 [DOI] [PubMed] [Google Scholar]
- Pata M. O., Hannun Y. A., Ng C. K. (2010). Plant sphingolipids: decoding the enigma of the Sphinx. New Phytol. 185, 611–630 10.1111/j.1469-8137.2009.03123.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer M., Bach M., Mueller M. J., Waller F. (2011). Free sphingobases induce RBOHD-dependent reactive oxygen species production in Arabidopsis leaves. FEBS Lett. 585, 3006–3010 10.1016/j.febslet.2011.08.016 [DOI] [PubMed] [Google Scholar]
- Peters C., Li M., Narasimhan R., Roth M., Welti R., Wang X. (2010). Nonspecific phospholipase C NPC4 promotes responses to abscisic acid and tolerance to hyperosmotic stress in Arabidopsis. Plant Cell 22, 2642–2659 10.1105/tpc.109.071720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C., Wang C., Wang X. (2002). Kinetic analysis of Arabidopsis phospholipase Dδ. Substrate preference and mechanism of activation by Ca2+ and phosphatidylinositol 4,5-biphosphate. J. Biol. Chem. 277, 49685–49690 10.1074/jbc.M200797200 [DOI] [PubMed] [Google Scholar]
- Qin W., Pappan K., Wang X. (1997). Molecular heterogeneity of phospholipase D (PLD). Cloning of PLDγ and regulation of plant PLDγ, -β, and -α by polyphosphoinositides and calcium. J. Biol. Chem. 272, 28267–28273 10.1074/jbc.272.40.25210 [DOI] [PubMed] [Google Scholar]
- Ryu S. B., Wang X. (1998). Increase in free linolenic and linoleic acids associated with phospholipase D-mediated hydrolysis of phospholipids in wounded castor bean leaves. Biochim. Biophys. Acta 1393, 193–202 [DOI] [PubMed] [Google Scholar]
- Sang Y., Zheng S., Li W., Huang B., Wang X. (2001a). Regulation of plant water loss by manipulating the expression of phospholipase Dα. Plant J. 28, 135–144 10.1046/j.1365-313X.2001.01138.x [DOI] [PubMed] [Google Scholar]
- Sang Y., Cui D., Wang X. (2001b). Phospholipase D and phosphatidic acid-mediated generation of superoxide in Arabidopsis. Plant Physiol. 126, 1449–1458 10.1104/pp.126.4.1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saucedo-Garcia M., Guevara-Garcia A., Gonzalez-Solis A., Cruz-Garcia F., Vazquez-Santana S., Markham J. E., Lozano-Rosas M. G., Dietrich C. R., Ramos-Vega M., Cahoon E. B., Gavilanes-Ruiz M. (2011). MPK6, sphinganine and the LCB2a gene from serine palmitoyltransferase are required in the signaling pathway that mediates cell death induced by long chain bases in Arabidopsis. New Phytol. 191, 943–957 10.1111/j.1469-8137.2011.03727.x [DOI] [PubMed] [Google Scholar]
- Shi L., Bielawski J., Mu J., Dong H., Teng C., Zhang J., Yang X., Tomishige N., Hanada K., Hannun Y. A., Zuo J. (2007). Involvement of sphingoid bases in mediating reactive oxygen intermediate production and programmed cell death in Arabidopsis. Cell Res. 17, 1030–1040 10.1038/cr.2007.100 [DOI] [PubMed] [Google Scholar]
- Strub G. M., Maceyka M., Hait N. C., Milstien S., Spiegel S. (2010). Extracellular and intracellular actions of sphingosine-1-phosphate. Adv. Exp. Med. Biol. 688, 141–155 10.1007/978-1-4419-6741-1_10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testerink C., Dekker H. L., Lim Z. Y., Johns M. K., Holmes A. B., Koster C. G., Ktistakis N. T., Munnik T. (2004). Isolation and identification of phosphatidic acid targets from plants. Plant J. 39, 527–536 10.1111/j.1365-313X.2004.02152.x [DOI] [PubMed] [Google Scholar]
- Testerink C., Larsen P. B., Van Der Does D., Van Himbergen J. A., Munnik T. (2007). Phosphatidic acid binds to and inhibits the activity of Arabidopsis CTR1. J. Exp. Bot. 58, 3905–3914 10.1093/jxb/erm243 [DOI] [PubMed] [Google Scholar]
- Umezawa T., Nakashima K., Miyakawa T., Kuromori T., Tanokura M., Shinozaki K., Yamaguchi-Shinozaki K. (2010). Molecular basis of the core regulatory network in ABA responses: sensing, signaling and transport. Plant Cell Physiol. 51, 1821–1839 10.1093/pcp/pcq156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Wang X. (2001). A novel phospholipase D of Arabidopsis that is activated by oleic acid and associated with the plasma membrane. Plant Physiol. 127, 1102–1112 10.1104/pp.127.1.345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Yang X., Tangchaiburana S., Ndeh R., Markham J. E., Tsegaye Y., Dunn T. M., Wang G. L., Bellizzi M., Parsons J. F., Morrissey D., Bravo J. E., Lynch D. V., Xiao S. (2008). An inositolphosphorylceramide synthase is involved in regulation of plant programmed cell death associated with defense in Arabidopsis. Plant Cell 20, 3163–3179 10.1105/tpc.108.060053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. (2001). Plant phospholipases. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 211–231 10.1146/annurev.arplant.52.1.211 [DOI] [PubMed] [Google Scholar]
- Wang X., Devaiah S. P., Zhang W., Welti R. (2006). Signaling functions of phosphatidic acid. Prog. Lipid Res. 45, 250–278 10.1016/j.plipres.2006.01.005 [DOI] [PubMed] [Google Scholar]
- Wang X., Xu L., Zheng L. (1994). Cloning and expression of phosphatidylcholine- hydrolyzing phospholipase D from Ricinus communis L. J. Biol. Chem. 269, 20312–20317 [PubMed] [Google Scholar]
- Worrall D., Liang Y. K., Alvarez S., Holroyd G. H., Spiegel S., Panagopulos M., Gray J. E., Hetherington A. M. (2008). Involvement of sphingosine kinase in plant cell signalling. Plant J. 56, 64–72 10.1111/j.1365-313X.2008.03579.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worrall D., Ng C. K., Hetherington A. M. (2003). Sphingolipids, new players in plant signaling. Trends Plant Sci. 8, 317–320 10.1016/S1360-1385(03)00128-6 [DOI] [PubMed] [Google Scholar]
- Yu L., Nie J., Cao C., Jin Y., Yan M., Wang F., Liu J., Xiao Y., Liang Y., Zhang W. (2010). Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol. 188, 762–773 10.1111/j.1469-8137.2010.03422.x [DOI] [PubMed] [Google Scholar]
- Zhang W., Qin C., Zhao J., Wang X. (2004). Phospholipase Dα1-derived phosphatidic acid interacts with ABI1 phosphatase 2C and regulates abscisic acid signaling. Proc. Natl. Acad. Sci. U.S.A. 101, 9508–9513 10.1073/pnas.0401167101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Wang C., Qin C., Wood T., Olafsdottir G., Welti R., Wang X. (2003). The oleate-stimulated phospholipase D, PLDδ, and phosphatidic acid decrease H2O2-induced cell death in Arabidopsis. Plant Cell 15, 2285–2295 10.1105/tpc.013961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhu H., Zhang Q., Li M., Yan M., Wang R., Wang L., Welti R., Zhang W., Wang X. (2009). Phospholipase Dα1 and phosphatidic acid regulate NADPH oxidase activity and production of reactive oxygen species in ABA-mediated stomatal closure in Arabidopsis. Plant Cell 21, 2357–2377 10.1105/tpc.108.061457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Wang X. (2004). Arabidopsis phospholipase Dα1 interacts with the heterotrimeric G-protein alpha-subunit through a motif analogous to the DRY motif in G-protein-coupled receptors. J. Biol. Chem. 279, 1794–1800 10.1074/jbc.C400473200 [DOI] [PubMed] [Google Scholar]
- Zheng L., Shan J., Krishnamoorthi R., Wang X. (2002). Activation of plant phospholipase Dα by phosphatidylinositol 4,5-bisphosphate: characterization of binding site and mode of action. Biochemistry 41, 4546–4553 10.1021/bi025841s [DOI] [PubMed] [Google Scholar]