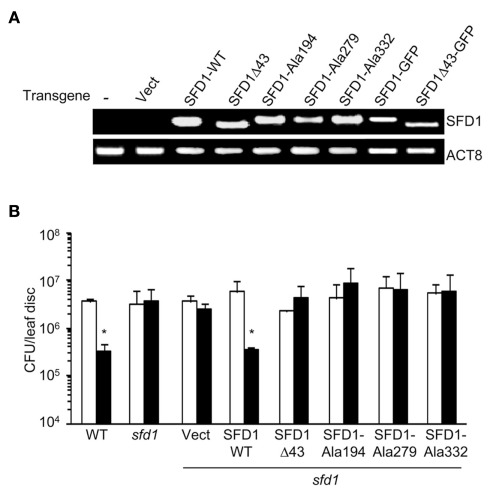
Figure 4.
Systemic acquired resistance in sfd1-1 transgenic plants expressing the wild type and mutant SFD1. (A) RT-PCR analysis of the SFD1 transgene expression in leaves of the sfd1-1 mutant plants transformed with the empty pMDC32 vector (Vect), the wild type (SFD1-WT) and the SFD1Δ43, SFD1–Ala194, SFD1–Ala279, and SFD1–Ala332 mutant constructs, and the SFD1–GFP and SFD1Δ43–GFP protein fusion expressing constructs. RNA extracted from non-transformed sfd1-1 mutant provided the negative control (−). The attR1-F plus a reverse primer (SFD-RCt) specific for the C-terminus of SFD1 were used for the PCR reaction to monitor expression of the SFD1 transgenes. This primer pair does not detect expression of the genomic SFD1. Expression of the ACTIN gene, ACT8, provided the control for RT-PCR. (B) SAR conferred resistance against P. syringae pv. maculicola in the wild type (WT) Arabidopsis accession Nössen plant, the sfd1-1 mutant, and sfd1-1 mutant plants transformed with the indicated constructs. SAR was induced by prior inoculation of a lower leaf with P. syringae pv. tomato DC3000 carrying the avrRpt2 avirulence gene (black bars). Plants similarly treated with 10 mM MgCl2 (white bars) provided the mock control for SAR. Three days after the primary inoculation the distal leaves were challenged with P. syringae pv. maculicola (Pma). Pma numbers were monitored 3 days post inoculation. Each bar represents the average colony forming units (CFU) of Pma ± SD in three samples each containing five leaf disks. An *indicates significant (P < 0.05) differences in bacterial numbers relative to the corresponding mock control.