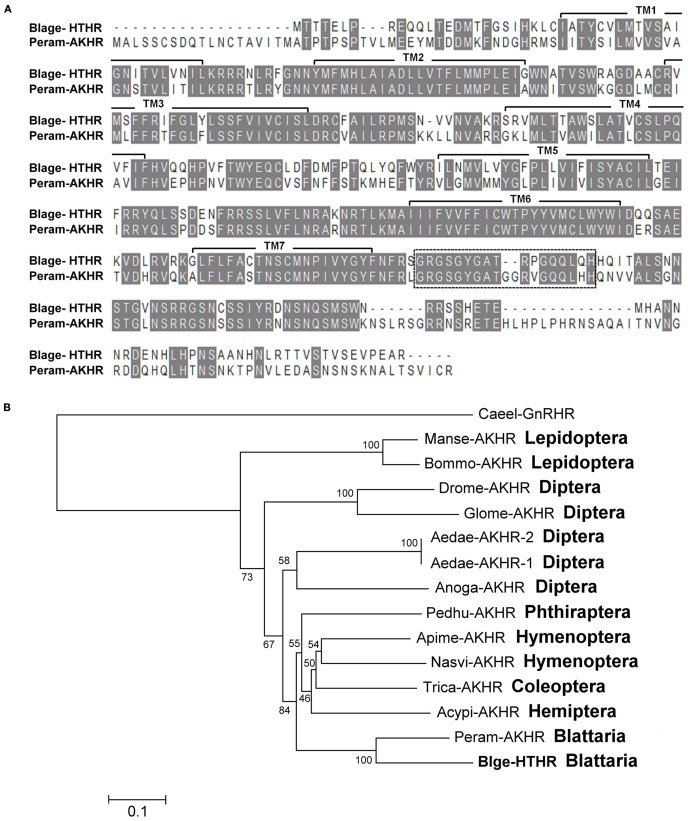
Figure 1.
(A) Alignment of the deduced amino acid sequences of the Blattella germanica HTH receptor (Blage-HTHR, GenBank GU591493) with the Periplaneta americana AKH receptor (Peram-AKHR, GenBank ABB20590). Identical amino acid residues are highlighted in gray. The seven transmembrane α-helices are indicated by TM1-7. The dotted-line rectangle indicates an insertion present in both cockroach species. (B) Neighbor joining tree of AKH receptor insect sequences. The tree was generated using MEGA 5 with 1000 bootstrap replicates. The evolutionary distance is given in units of the number of amino acid substitutions per site. The abbreviated names of insect AKHR are: Acypi-AKHR (Acyrthosiphon pisum), Aedae-AKHR-1 and Aedae-AKHR2 (Aedes aegypti), Anoga-AKHR (Anopheles gambiae), Apime-AKHR (Apis mellifera), Blage-HTHR (Blattella germanica), Bommo-AKHR (Bombyx mori), Drome-AKHR (Drosophila melanogaster), Glomo-AKHR (Glossina morsitans), Manse-AKHR (Manduca sexta), Nasvi-AKHR (Nasonia vitripennis), Pedhu-AKHR (Pediculus humanus), Peram-AKHR (Periplaneta americana), and Trica-AKHR (Tribolium castaneum). Gonadotropin-releasing hormone receptor of Caenorhabditis elegans (Caeel-GnRHR) was used as outgroup.