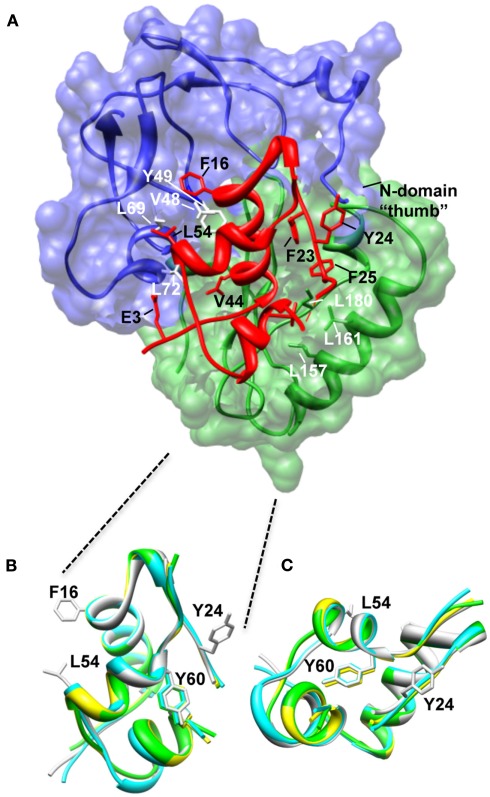
Figure 2.
Structure of the N-BP-4:IGF-I:C-BP-4 complex (PDB 2DSR; Sitar et al., 2006). (A) The N-domain of BP-4 (blue, residues 3–82) contacts the C-domain of BP-4 (green, residues 151–232) to form a high-affinity complex with IGF-I (red). IGF-I residues Phe16 and Leu54 insert into a cleft in the N-BP-4 encompassing residues Val48, Tyr49, Leu69, and Leu72 (Kalus et al., 1998). The “thumb” region of the N-domain and residues Leu157, Leu161, and Ile180 of the IGFBP-4 C-domain are in contact with IGF-I residues Phe23, Tyr24, Phe25, and Val44, which are involved in IGF-1R binding, resulting in steric hindrance of IGF-I binding to the receptor (PDB 2DSR, Sitar et al., 2006). (B,C) Superposition of IGF-I crystal structures highlighting that the Tyr60 side chain remains in the same position whether IGF-I is bound to IGFBPs or free. Ribbon views of IGF-I in the free form (bound to detergent molecules) PDB 1GZR (green; Brzozowski et al., 2002) and PDB 1IMX (yellow; Vajdos et al., 2001), the IGF-I‚ mini-N-BP-5 binary complex (PDB 1H59, blue; Zeslawski et al., 2001), and the IGF-I‚ N-BP-4‚ C-BP-4 ternary complex (PDB 2DSR, light gray; Sitar et al., 2006) are superimposed over backbone heavy atoms of the three helices (Ala8–Cys18, Gly42–Cys48, Leu54–Cys61). The side chains of Phe16, Phe49, and Leu54 are shown. The structures are shown in two different orientations (B,C). The structures in (B) are in an orientation equivalent to that of the IGF-I structure in (A). Parts of the flexible regions (residues 1–2, 26–42, and 63–70) are excluded for clarity. Molecular graphic images were produced with the UCSF Chimera program from Resource for Biocomputing, Visualization, and Informatics at the University of California at San Francisco.