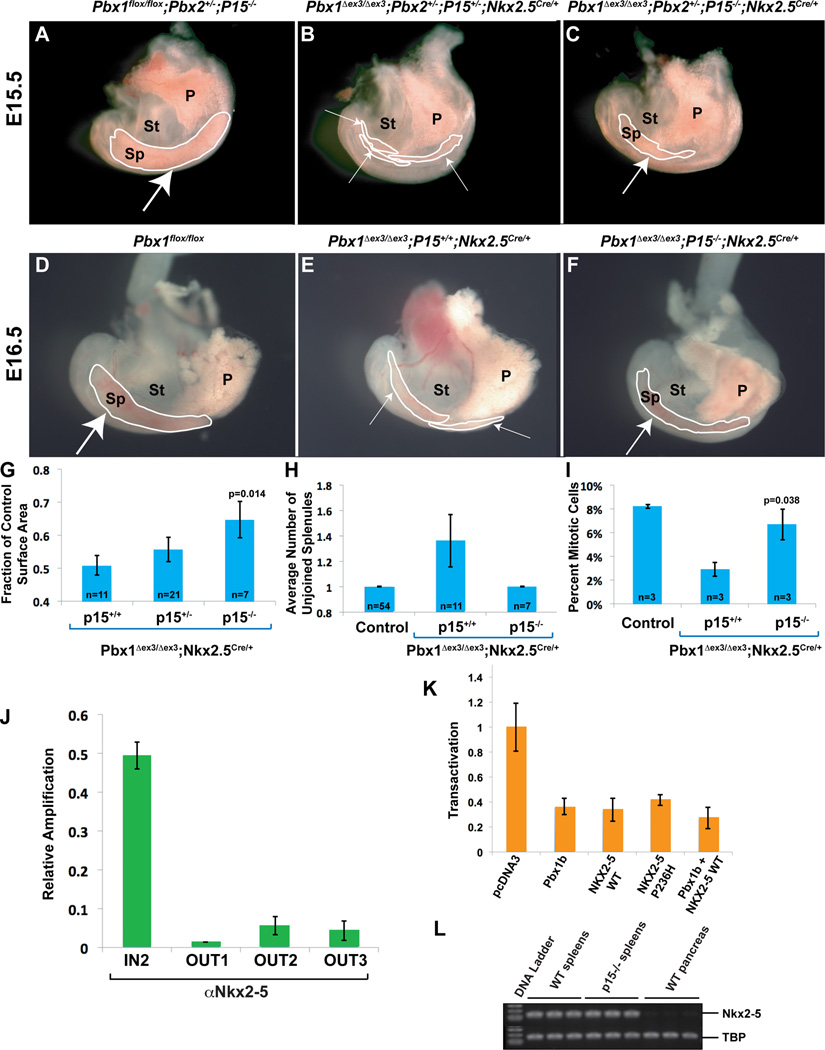
Figure 5. Genetic ablation of p15Ink4b, which is bound by Nkx2-5 in its cis-regulatory elements, partially rescues the spleen phenotypes.
(A–F) Partial rescue of spleen size and morphogenesis in E15.5 (A–C) and E16.5 (D–F) Pbx1Δex3/Δex3;Nkx2-5Cre/+ mutants on a p15-deficient background. Embryonic spleens (outlined in white) in which Pbx1 has been deleted in the mesenchyme either alone (E,F) or with Pbx2 (B,C), show a significant increase in spleen size and rescued morphology on a p15-deficient background (medium white arrows; C,F), versus littermates which retain p15 (thin white arrows; B,E). Control spleens (thick white arrows; A,D) for comparison. (G–I) Summary of genetic rescue data at E16.5. (G) Spleen surface area measurements (Experimental Procedures) normalized to controls reveal that p15-null Pbx1 mutant spleens are significantly larger than p15+/+ mutant spleens (p=0.014). (H) All rescued embryos develop one single spleen anlage. (I) Proliferation rate in p15−/− mutant spleen mesenchyme is restored to near WT levels, significantly higher than in p15+/+ Pbx1 mutants (p=0.038). Number of samples (n) indicated on corresponding bar of each graph. P, pancreas; Sp, spleen; St, stomach. (J) ChIP shows Nkx2-5 binding to the p15 promoter. Primers located within the p15 promoter (p15Ink4b IN2), but not non-specific primers (OUT1, OUT2, and OUT3), amplify a specific product from αNkx2-5-immunoprecipitated chromatin from SPCL2 cells, as assessed by qPCR and normalized to amplification of input chromatin. (K) Transient transfection of NIH 3T3 cells with 800ng of Pbx1b, NKX2-5 WT or NKX2-5 P236H, or co-transfection of Pbx1b and NKX2-5 in combination (500ng each), causes a reduction in p15 promoter activity. Transactivation normalized to empty expression vector (pcDNA3). Data are mean ± SEM of one out of three independent experiments performed in triplicate. (L) Semi-qRT-PCR on E16.5 mouse splenic RNA does not reveal perturbations of Nkx2-5 in p15−/− mutants versus controls. E16.5 WT pancreas RNA used as negative control for Nkx2-5 expression. All data are mean ± SEM. TBP, TATA-binding protein. See also Figure S6.