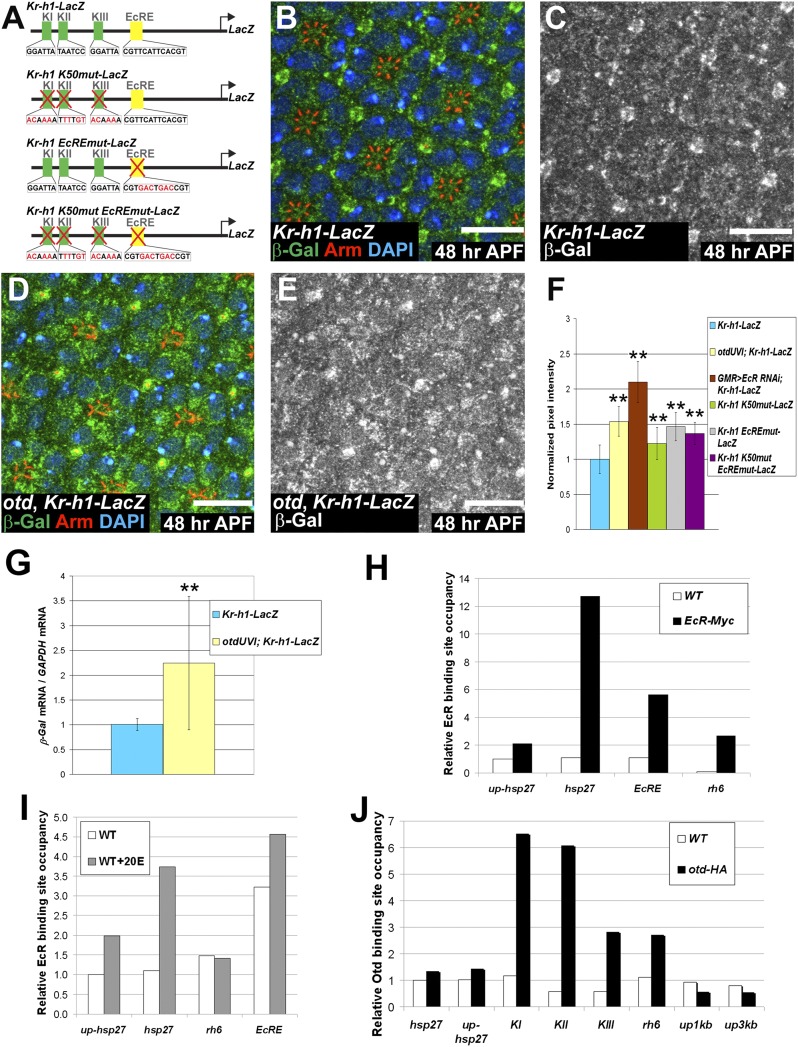
Fig. 4.
Both Otd and EcR repress the transcription of Kr-h1. (A) Reporter genes generated to monitor Kr-h1 regulation. KI–KIII represent the three K50 sites (green boxes) and EcRE represents the 20E receptor binding site (yellow box). Mutations and modified sequences are highlighted in red. (B–E) 48 h APF. (Scale bars, 10 μm.) Kr-h1-LacZ expressed (B and C) in a WT and (D and E) in an otdUVI background. (B and D) β-Gal (green), Arm (red), and DAPI (blue). (C and E) β-Gal (gray). (F) Quantification of pixel intensity of the β-Gal channel. WT ommatidia (n = 192) (blue), otdUVI ommatidia (n = 148) (yellow), GMR-GAL4/UAS-EcR RNAi (n = 52) (brown), WT ommatidia expressing Kr-h1 K50mut-LacZ, (n = 122) (green), WT ommatidia expressing Krh1 EcREmut-LacZ, (n = 132) (gray), and WT ommatidia expressing Krh1 K50mut EcREmut-LacZ, (n = 77) (purple). Asterisks indicate statistically significant differences with WT ommatidia expressing Kr-h1-LacZ, P ≤ 10−17. Quantifications were performed in males. (G) qPCR quantification of LacZ mRNA normalized to GAPDH mRNA levels comparing Kr-h1-LacZ retinas (blue) and otdUVI; Kr-h1-LacZ retinas (yellow) at 48 h APF. n = 9 independent mRNA extracts from otd and WT retinas, P ≤ 0.04. Error bars represent SD. (H–J) Representative experiment of n ≥ 2 (H) ChIP carried out using an anti-MYC antibody on WT (white) and EcR-MYC-overexpressing cells (black). (I) ChIP carried out using an anti-EcR antibody on nontreated (white) and 20E treated WT cells (gray). (J) ChIP carried out using an anti-HA antibody on WT (white) and otd-HA-overexpressing cells (black).