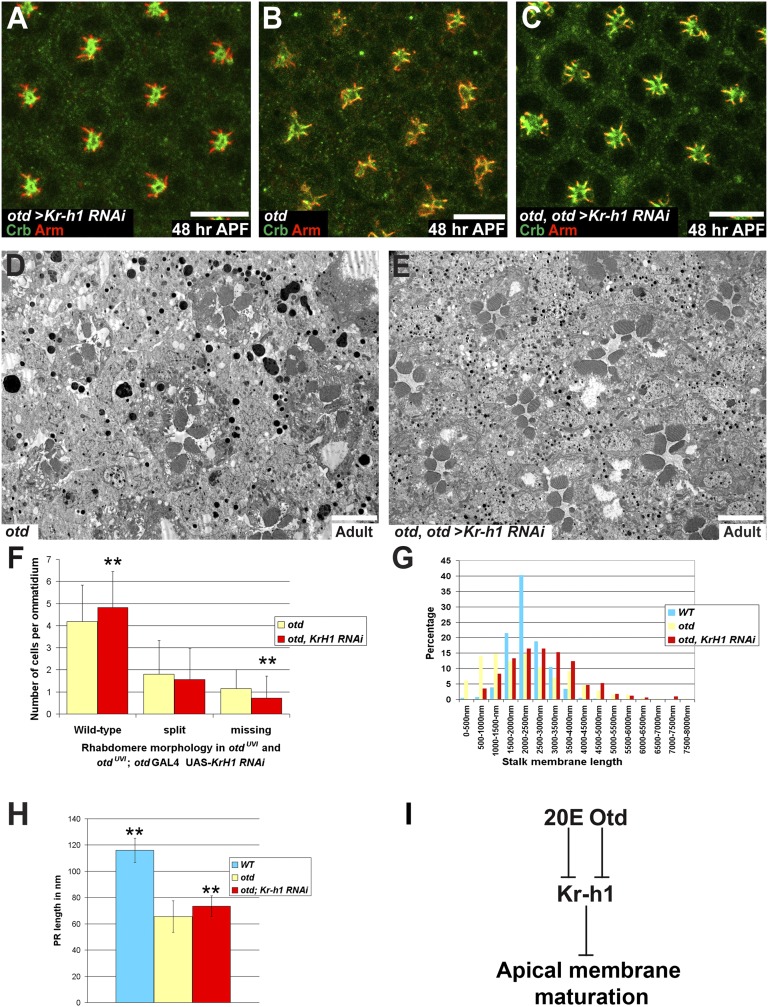
Fig. 6.
Knocking down Kr-h1 in an otd background partially suppresses PR maturation defects. (A–C) Crb (green), Arm (red), ommatidia at 48 h APF. (Scale bars, 10 μm.) (A) otd-Gal4/UAS-Kr-h1 RNAi. (B) otdUVI/Y; otd-Gal4/SM6:TM6 PRs. (C) otdUVI/Y; otd-Gal4/UAS-Kr-h1 RNAi. (D and E) (Scale bars, 5 μm.) (D) otdUVI/Y; otd-Gal4/SM6:TM6 and (E) otdUVI/Y; otd-Gal4/UAS-Kr-h1 RNAi. (F) Rhabdomere phenotype in otdUVI/Y; otd-Gal4/SM6:TM6 (yellow) and in an otdUVI/Y; otd-Gal4/UAS-Kr-h1 RNAi background (red). Asterisks indicate a statistically significant difference. n = 1,122 otd PRs and 816 otd + Kr-h1 RNAi PRs, P ≤ 10−3. (G) Stalk membrane length in WT PRs (blue), in otdUVI/Y; otd-Gal4/SM6:TM6 (yellow), and in otdUVI/Y; otd-Gal4/UAS-Kr-h1 RNAi PRs (red). (H) PR length over several Z-sections of WT PRs (blue), in otdUVI/Y; otd-Gal4/SM6:TM6 (yellow), and in otdUVI/Y; otd-Gal4/UAS-Kr-h1 RNAi PRs (red). Asterisks indicate a statistically significant difference. n = 30 PRs for each genotype P ≤ 0.0055. (I) Working model.