Abstract
Recent technical advances have begun to realize the potential of molecular beacons to test for diverse infections in clinical diagnostic laboratories. These include the ability to test for, and quantify, multiple pathogens in the same clinical sample, and to detect antibiotic resistant strains within hours. The design principles of molecular beacons have also spawned a variety of allied technologies.
Introduction
Imagine a magic reagent to which is added a droplet of body fluid from a patient. A glow appears in the tube and the color of the glow identifies the pathogen responsible for the patient's illness. Such was the promise of molecular beacons when they were first introduced 15 years ago [1]. That promise is now bearing fruit, as powerful new tests for diverse infections are being introduced into clinical diagnostic laboratories.
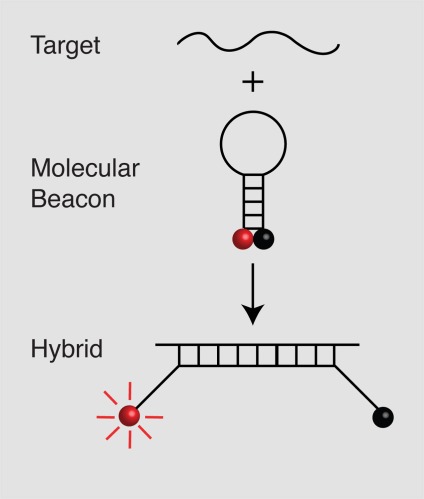
Molecular beacons are hairpin-shaped oligonucleotide probes that become fluorescent upon hybridization to an RNA or DNA target sequence. Their loops serve as probes and are about 15 to 25 nucleotides long. Their stems serve to bring the two ends of the molecule, which are linked to a fluorophore and a quencher, into close proximity. Although the stems are only 5 to 7 nucleotides long, they keep the labels in close proximity so that the fluorescence of the fluorophore is quenched in the free probes. However, upon binding to their target, they undergo a spontaneous conformational reorganization that removes the fluorophore from the vicinity of the quencher and restores its fluorescence (Figure 1). Molecular beacons specific for different target sequences and labeled with differently colored fluorophores can be used together to simultaneously identify and quantitate multiple targets in the same assay tube [2].
Figure 1. Principle of operation of molecular beacons.
In the free probes, a fluorophore attached at one of the molecule (red ball) is held close to a quencher (black ball) attached at the other end of the molecule, so that the probe is not fluorescent. The interaction of the sequence in the loop with the target separates the fluorophore and quencher and causes the fluorophore to become fluorescent.
Given this fluorogenic response, the addition of the probes to nucleic acids isolated from a clinical sample containing a relatively large number of infectious agents would be sufficient to generate a detectable signal. However, human clinical samples contain only miniscule amounts of pathogen-derived nucleic acids. It is therefore necessary to first amplify an identifying target sequence from the pathogen's nucleic acid. In a typical assay, nucleic acids are extracted from the sample, and a segment of the pathogen's genome (containing the target sequence) is amplified in a polymerase chain reaction (PCR), or in a nucleic acid sequence-based amplification (NASBA) reaction, while simultaneously monitoring the fluorescence of the target-specific molecular beacons in the reaction mixture [1-3]. Since fluorescence is measured in sealed tubes, carryover contamination, which plagued earlier assays, cannot occur. Moreover, the measurement of fluorescence is in real time, as opposed to making measurements at end points, which enables quantitation over an extremely wide dynamic range of target concentrations [4].
Recent advances
Soon after the introduction of molecular beacons and other target-specific fluorogenic probes, such as TaqMan probes [1,5], spectrofluorometric thermal cyclers became available that could perform PCR assays while simultaneously monitoring fluorescence changes. In addition, automated nucleic acid extraction protocols were introduced. With the availability of these resources, the development of nucleic acid-based assays for pathogens became relatively simple, and there are now a large number of tests available for the detection of human pathogens, and food and environmental contaminants (Table 1).
Table 1. Molecular beacon based assays for pathogens.
| Organism | Amplification Scheme | References |
|---|---|---|
| Human Pathogens | ||
| HIV | NASBA, PCR | [4,19,20] |
| HTLV | PCR | [4,21] |
| Oncogenic HPV | NASBA, PCR | [22-24] |
| Mycobacterium tuberculosis | PCR | [9-11,25] |
| hMPV | NASBA | [26] |
| RSV | NASBA, PCR | [27-29] |
| Enterovirus | NASBA | [30,31] |
| Influenza virus | NASBA, PCR | [32,33] |
| Entamoeba histolytica | PCR | [34] |
| Methicillin-resistant Staphylococcus | PCR | [35-37] |
| Hepatitis B | NASBA, PCR | [38,39] |
| HPIV | NASBA | [40] |
| West Nile virus | NASBA, PCR | [41,42] |
| Candida dubliniensis | PCR | [43] |
| Scedosporium | PCR | [44] |
| Pan-bacteria | NASBA, PCR, melting analysis | [6,7] |
| Pan-fungi | NASBA, PCR | [6] |
| 27 Mycobacterial species | PCR, melting analysis | [8] |
| 6 Pneumonia-causing agents | PCR | [45] |
| Chlamydophila pneumoniae | NASBA | [46] |
| Multiple bacteria in blood stream | PCR | [47] |
| Lyme disease spirochetes | PCR | [48] |
| Plasmodium | NASBA, PCR | [49,50] |
| Food Pathogens | ||
| Salmonella | PCR | [51] |
| Escherichia coli | NASBA | [52] |
| Listeria | NASBA | [53] |
| 8 food pathogens | PCR, melting analysis | [54] |
| Environmental Pathogens | ||
| Salmonella | PCR | [55] |
| Escherichia coli | PCR | [56] |
| Baylisascaris procyonis | PCR | [57] |
| Vibrio cholerae | NASBA, PCR | [58,59] |
| Bacillus anthracis | PCR | [60] |
Abbreviations: hMPV, human metapneumovirus; HPIV, human parainfluenza virus; RSV, respiratory syncytial virus.
Most of the assays listed in the table are designed to detect an individual pathogenic species. However, it is often desirable to determine the type of pathogens that are present in a clinical sample, for example, does the sample contain Gram-negative bacteria, Gram-positive bacteria, or a fungal species [6]? These tests would be even more powerful if they could determine which species (from a selected list) is present, and indeed, investigators have recently developed tests that do this. For example, in a single assay, Chakravorty and his colleagues [7] were able to identify which pathogenic or commensal bacterial species was present in a blood sample from a list of 111 species in 64 different genera. Similarly, El-Hajj and her colleagues [8] have developed an assay that distinguishes 27 different species of Mycobacteria. These assays include several molecular beacons labeled with differently colored fluorophores in the same tube. Current instruments are able to distinguish up to seven fluorescent colors in the same tube, which suggests a limit of seven targets. So, to achieve this high level of multiplexing, the assays not only distinguish the fluorescence of each differently colored molecular beacon, they measure the characteristic stability of the probe-target hybrids that each molecular beacon forms by raising the temperature to see when each molecular beacon dissociates from the target (causing a marked decrease in fluorescence). This analysis identifies the target by indicating the relationship of the probe sequence to the target sequence. If the probe is a good match to the target, the probe-target hybrid is quite stable, and this stability is reflected in a high melting temperature, whereas, if the probe is not a good match to the target, the probe-target hybrid is less stable and melts apart at a lower temperature.
Some of these tests can also determine whether the specific pathogen has acquired drug resistance. For example, an assay developed by Alland and colleagues [9-11] for Mycobacterium tuberculosis detects the bacterial RNA polymerase gene with several different molecular beacons that are so specific that the presence of any mutation in the target region causes one of the differently colored molecular beacons not to bind to the target (indicating that the commonly used antibiotic rifampicin will not be able to inhibit the activity of the encoded RNA polymerase). This PCR assay takes only two hours, whereas traditional culture-based tests take at least two weeks.
In a number of clinical situations, it is necessary to detect hundreds or thousands of targets at the same time. For example, such situations occur when the mutations responsible for drug resistance are scattered over long sequence stretches, or can be present on multiple genes. To probe for a large number of mutations, new assay formats are under development that simultaneously screen genomes with thousands of molecular beacons. In these formats, different molecular beacons are immobilized at different locations of a planner array [12], on the surface of beads trapped in microfluidic chambers [13], or on different locations of nanowires [14]. The locations that become fluorescent indicate which mutations are present.
It is not always necessary to amplify nucleic acid to detect a pathogen. Molecular beacons complementary to species-specific regions of ribosomal RNA can be used to identify bacterial and fungal pathogens by in situ hybridization. In novel “molecular blood culture” assays, pathogens are grown for a short period and then identified by in situ hybridization with molecular beacons, followed by imaging [15].
In addition to their main attribute (that they generate signals without having to separate probe target-hybrids from excess probes, thereby enabling sealed-tube assays), molecular beacons also introduced several design principles that have inspired the development of other kinds of probes. Among these are the use of non-fluorescent dyes as quenchers of fluorescence, the use of hairpins as a means of keeping fluorophores in close contact with quenchers, and the use of hairpins to increase the specificity of hybridization. A small industry of non-fluorescent quenchers, often called “dark” quenchers, with names such as Eclipse, Iowa Black, BlackBerry, and Black Hole Quenchers has been spawned [16]. Dark quenchers afford a larger extent of multiplexing, since they do not crowd the available spectrum by emitting light. A number of novel probe formats that utilize hairpins as their central element have also been introduced. For example, Amplifluor Primers contain a 5’ hairpin that possesses a fluorophore and a quencher in close contact, but once the resulting amplicon (a piece of DNA that has been synthesized using amplification techniques) is copied, the fluorophore and quencher are separated, generating a bright fluorescence signal [17]. In another variant, called Scorpion Primers, a molecular beacon is attached at the 5’ end of the primer, but in this case, it cannot be copied. However, the molecular beacon is designed to bind to a segment of the amplified DNA. Since the probe and the target are now in the same molecule, the molecular beacon can come around and bind to the target. This intramolecular interaction happens more readily than the intermolecular binding of a normal molecular beacon [18].
Future directions
Recent years have seen a wave of technical advances that have begun to realize the potential of molecular beacons to test for diverse human pathogens in clinical diagnostic laboratories, and also food and environmental contaminants. These include new techniques with the ability to test for, and quantify, multiple pathogens in the same clinical sample, and to detect antibiotic resistant strains within hours.
As molecular beacons and allied technologies are adopted in the clinical diagnostics industry, physicians will be able to diagnose diseases with greater precision than ever before.
Acknowledgments
This work is supported by the National Institutes of Mental Health Grant 079197.
Abbreviations
- NASBA
nucleic acid sequence-based amplification
- PCR
polymerase chain reaction
Competing Interests
The authors are inventors of molecular beacons technology, which is patented and licensed non-exclusively to more than 70 licensees, and the authors receive a portion of the resulting licensing income, both personally, and as unrestricted funds that support ongoing research in their laboratory
The electronic version of this article is the complete one and can be found at: http://f1000.com/reports/m/4/10
References
- 1.Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat Biotechnol. 1996;14:303–8. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 2.Tyagi S, Bratu DP, Kramer FR. Multicolor molecular beacons for allele discrimination. Nat Biotechnol. 1998;16:49–53. doi: 10.1038/nbt0198-49. [DOI] [PubMed] [Google Scholar]
- 3.Leone G, van Schijndel H, van Gemen B, Kramer FR, Schoen CD. Molecular beacon probes combined with amplification by NASBA enable homogeneous, real-time detection of RNA. Nucleic Acids Res. 1998;26:2150–5. doi: 10.1093/nar/26.9.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vet JA, Majithia AR, Marras SA, Tyagi S, Dube S, Poiesz BJ, Kramer FR. Multiplex detection of four pathogenic retroviruses using molecular beacons. Proc Natl Acad Sci U S A. 1999;96:6394–9. doi: 10.1073/pnas.96.11.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6:986–94. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 6.Zhao Y, Park S, Kreiswirth BN, Ginocchio CC, Veyret R, Laayoun A, Troesch A, Perlin DS. Rapid real-time nucleic acid sequence-based amplification-molecular beacon platform to detect fungal and bacterial bloodstream infections. J Clin Microbiol. 2009;47:2067–78. doi: 10.1128/JCM.02230-08. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Sanjay Tyaji 23 Apr 2012
- 7.Chakravorty S, Aladegbami B, Burday M, Levi M, Marras SA, Shah D, El-Hajj HH, Kramer FR, Alland D. Rapid universal identification of bacterial pathogens from clinical cultures by using a novel sloppy molecular beacon melting temperature signature technique. J Clin Microbiol. 2010;48:258–67. doi: 10.1128/JCM.01725-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El-Hajj HH, Marras SA, Tyagi S, Shashkina E, Kamboj M, Kiehn TE, Glickman MS, Kramer FR, Alland D. Use of sloppy molecular beacon probes for identification of mycobacterial species. J Clin Microbiol. 2009;47:1190–8. doi: 10.1128/JCM.02043-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boehme CC, Nabeta P, Hillemann D, Nicol MP, Shenai S, Krapp F, Allen J, Tahirli R, Blakemore R, Rustomjee R, Milovic A, Jones M, O'Brien SM, Persing DH, Ruesch-Gerdes S, Gotuzzo E, Rodrigues C, Alland D, Perkins MD. Rapid molecular detection of tuberculosis and rifampin resistance. N Engl J Med. 2010;363:1005–15. doi: 10.1056/NEJMoa0907847. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 9Evaluated by Anthony Harries12 Nov 2010, Elizabeth Molyneux 13 Oct 2010
- 10.El-Hajj HH, Marras SA, Tyagi S, Kramer FR, Alland D. Detection of rifampin resistance in Mycobacterium tuberculosis in a single tube with molecular beacons. J Clin Microbiol. 2001;39:4131–7. doi: 10.1056/NEJMoa0907847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Piatek AS, Tyagi S, Pol AC, Telenti A, Miller LP, Kramer FR, Alland D. Molecular beacon sequence analysis for detecting drug resistance in Mycobacterium tuberculosis. Nat Biotechnol. 1998;16:359–63. doi: 10.1038/nbt0498-359. [DOI] [PubMed] [Google Scholar]
- 12.Situma C, Moehring AJ, Noor MA, Soper SA. Immobilized molecular beacons: a new strategy using UV-activated poly(methyl methacrylate) surfaces to provide large fluorescence sensitivities for reporting on molecular association events. Anal Biochem. 2007;363:35–45. doi: 10.1016/j.ab.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Sanjay Tyaji 23 Apr 2012
- 13.Zuo X, Yang X, Wang K, Tan W, Wen J. A novel sandwich assay with molecular beacon as report probe for nucleic acids detection on one-dimensional microfluidic beads array. Anal Chim Acta. 2007;587:9–13. doi: 10.1016/j.aca.2007.01.032. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Sanjay Tyaji 23 Apr 2012
- 14.Stoermer RL, Cederquist KB, McFarland SK, Sha MY, Penn SG, Keating CD. Coupling molecular beacons to barcoded metal nanowires for multiplexed, sealed chamber DNA bioassays. J Am Chem Soc. 2006;128:16892–903. doi: 10.1021/ja0658261. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Sanjay Tyaji 23 Apr 2012
- 15. http://miacom-diagnostics.com
- 16.Marras SA. Interactive fluorophore and quencher pairs for labeling fluorescent nucleic acid hybridization probes. Mol Biotechnol. 2008;38:247–55. doi: 10.1007/s12033-007-9012-9. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Sanjay Tyaji 23 Apr 2012
- 17.Nazarenko IA, Bhatnagar SK, Hohman RJ. A closed tube format for amplification and detection of DNA based on energy transfer. Nucleic Acids Res. 1997;25:2516–21. doi: 10.1093/nar/25.12.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whitcombe D, Theaker J, Guy SP, Brown T, Little S. Detection of PCR products using self-probing amplicons and fluorescence. Nat Biotechnol. 1999;17:804–7. doi: 10.1038/11751. [DOI] [PubMed] [Google Scholar]
- 19.McClernon DR, Vavro C, St Clair M. Evaluation of a real-time nucleic acid sequence-based amplification assay using molecular beacons for detection of human immunodeficiency virus type 1. J Clin Microbiol. 2006;44:2280–2. doi: 10.1128/JCM.02187-05. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Sanjay Tyaji 23 Apr 2012
- 20.Demetriou VL, van de Vijver DA, Kousiappa I, Balotta C, Clotet B, Grossman Z, Jørgensen LB, Lepej SZ, Levy I, Nielsen C, Paraskevis D, Poljak M, Roman F, Ruiz L, Schmidt JC, Vandamme AM, Van Laethem K, Vercauteren J, Kostrikis LG. Cellular HIV-1 DNA levels in drug sensitive strains are equivalent to those in drug resistant strains in newly-diagnosed patients in Europe. PLoS One. 2010;5:e10976. doi: 10.1371/journal.pone.0010976. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Sanjay Tyaji 23 Apr 2012
- 21.Besson G, Kazanji M. One-step, multiplex, real-time PCR assay with molecular beacon probes for simultaneous detection, differentiation, and quantification of human T-cell leukemia virus types 1, 2, and 3. J Clin Microbiol. 2009;47:1129–35. doi: 10.1128/JCM.02006-08. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Sanjay Tyaji 23 Apr 2012
- 22.Molden T, Kraus I, Skomedal H, Nordstrom T, Karlsen F. PreTect HPV-Proofer: real-time detection and typing of E6/E7 mRNA from carcinogenic human papillomaviruses. J Virol Methods. 2007;142:204–12. doi: 10.1016/j.jviromet.2007.01.036. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Sanjay Tyaji 23 Apr 2012
- 23.Ratnam S, Coutlee F, Fontaine D, Bentley J, Escott N, Ghatage P, Gadag V, Holloway G, Bartellas E, Kum N, Giede C, Lear A. Clinical performance of the PreTect HPV-Proofer E6/E7 mRNA assay in comparison with that of the Hybrid Capture 2 test for identification of women at risk of cervical cancer. J Clin Microbiol. 2010;48:2779–85. doi: 10.1128/JCM.00382-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takacs T, Jeney C, Kovacs L, Mozes J, Benczik M, Sebe A. Molecular beacon-based real-time PCR method for detection of 15 high-risk and 5 low-risk HPV types. J Virol Methods. 2008;149:153–62. doi: 10.1016/j.jviromet.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 25.Kumar P, Nath K, Rath B, Sen MK, Vishalakshi P, Chauhan DS, Katoch VM, Singh S, Tyagi S, Sreenivas V, Prasad HK. Visual format for detection of Mycobacterium tuberculosis and M. bovis in clinical samples using molecular beacons. J Mol Diagn. 2009;11:430–8. doi: 10.2353/jmoldx.2009.080135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ginocchio CC, Manji R, Lotlikar M, Zhang F. Clinical evaluation of NucliSENS magnetic extraction and NucliSENS analyte-specific reagents for real-time detection of human metapneumovirus in pediatric respiratory specimens. J Clin Microbiol. 2008;46:1274–80. doi: 10.1128/JCM.01567-07. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Sanjay Tyaji 23 Apr 2012
- 27.Manji R, Lotlikar M, Zhang F, Ginocchio CC. Clinical evaluation of NucliSENS magnetic extraction and NucliSENS analytical specific reagents for the real-time detection of respiratory syncytial virus (RSV) in paediatric respiratory specimens. J Clin Pathol. 2009;62:998–1002. doi: 10.1136/jcp.2009.066688. [DOI] [PubMed] [Google Scholar]
- 28.Deiman B, Schrover C, Moore C, Westmoreland D, van de Wiel P. Rapid and highly sensitive qualitative real-time assay for detection of respiratory syncytial virus A and B using NASBA and molecular beacon technology. J Virol Methods. 2007;146:29–35. doi: 10.1016/j.jviromet.2007.05.030. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Sanjay Tyaji 23 Apr 2012
- 29.O'Shea MK, Ryan MA, Hawksworth AW, Alsip BJ, Gray GC. Symptomatic respiratory syncytial virus infection in previously healthy young adults living in a crowded military environment. Clin Infect Dis. 2005;41:311–17. doi: 10.1086/431591. [DOI] [PubMed] [Google Scholar]
- 30.Landry ML, Garner R, Ferguson D. Real-time nucleic acid sequence-based amplification using molecular beacons for detection of enterovirus RNA in clinical specimens. J Clin Microbiol. 2005;43:3136–9. doi: 10.1128/JCM.43.7.3136-3139.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Capaul SE, Gorgievski-Hrisoho M. Detection of enterovirus RNA in cerebrospinal fluid (CSF) using NucliSens EasyQ Enterovirus assay. J Clin Virol. 2005;32:236–40. doi: 10.1016/j.jcv.2004.08.006. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Sanjay Tyaji 23 Apr 2012
- 32.Muradrasoli S, Mohamed N, Belak S, Czifra G, Herrmann B, Berencsi G, Blomberg J. Broadly targeted triplex real-time PCR detection of influenza A, B and C viruses based on the nucleoprotein gene and a novel “MegaBeacon” probe strategy. J Virol Methods. 2010;163:313–22. doi: 10.1016/j.jviromet.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Sanjay Tyaji 23 Apr 2012
- 33.Moore C, Hibbitts S, Owen N, Corden SA, Harrison G, Fox J, Gelder C, Westmoreland D. Development and evaluation of a real-time nucleic acid sequence based amplification assay for rapid detection of influenza A. J Med Virol. 2004;74:619–28. doi: 10.1002/jmv.20221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roy S, Kabir M, Mondal D, Ali IK, Petri WA, Jr., Haque R. Real-time-PCR assay for diagnosis of Entamoeba histolytica infection. J Clin Microbiol. 2005;43:2168–72. doi: 10.1128/JCM.43.5.2168-2172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Sanjay Tyaji 23 Apr 2012
- 35.Warren DK, Liao RS, Merz LR, Eveland M, Dunne WM., Jr. Detection of methicillin-resistant Staphylococcus aureus directly from nasal swab specimens by a real-time PCR assay. J Clin Microbiol. 2004;42:5578–81. doi: 10.1128/JCM.42.12.5578-5581.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Sanjay Tyaji 23 Apr 2012
- 36.Huletsky A, Giroux R, Rossbach V, Gagnon M, Vaillancourt M, Bernier M, Gagnon F, Truchon K, Bastien M, Picard FJ, van Belkum A, Ouellette M, Roy PH, Bergeron MG. New real-time PCR assay for rapid detection of methicillin-resistant Staphylococcus aureus directly from specimens containing a mixture of staphylococci. J Clin Microbiol. 2004;42:1875–84. doi: 10.1128/JCM.42.5.1875-1884.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sinsimer D, Leekha S, Park S, Marras SA, Koreen L, Willey B, Naidich S, Musser KA, Kreiswirth BN. Use of a multiplex molecular beacon platform for rapid detection of methicillin and vancomycin resistance in Staphylococcus aureus. J Clin Microbiol. 2005;43:4585–91. doi: 10.1128/JCM.43.9.4585-4591.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sum SS, Wong DK, Yuen MF, Yuan HJ, Yu J, Lai CL, Ho D, Zhang L. Real-time PCR assay using molecular beacon for quantitation of hepatitis B virus DNA. J Clin Microbiol. 2004;42:3438–40. doi: 10.1128/JCM.42.8.3438-3440.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yates S, Penning M, Goudsmit J, Frantzen I, van de Weijer B, van Strijp D, van Gemen B. Quantitative detection of hepatitis B virus DNA by real-time nucleic acid sequence-based amplification with molecular beacon detection. J Clin Microbiol. 2001;39:3656–65. doi: 10.1128/JCM.39.10.3656-3665.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Sanjay Tyaji 23 Apr 2012
- 40.Hibbitts S, Rahman A, John R, Westmoreland D, Fox JD. Development and evaluation of NucliSens basic kit NASBA for diagnosis of parainfluenza virus infection with 'end-point' and 'real-time' detection. J Virol Methods. 2003;108:145–55. doi: 10.1016/S0166-0934(02)00268-9. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Sanjay Tyaji 23 Apr 2012
- 41.Lanciotti RS, Kerst AJ. Nucleic acid sequence-based amplification assays for rapid detection of West Nile and St. Louis encephalitis viruses. J Clin Microbiol. 2001;39:4506–13. doi: 10.1128/JCM.39.12.4506-4513.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee DH, Mathew J, Pfahler W, Ma D, Valinsky J, Prince AM, Andrus L. Individual donor nucleic acid amplification testing for detection of West Nile virus. J Clin Microbiol. 2005;43:5111–6. doi: 10.1128/JCM.43.10.5111-5116.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Sanjay Tyaji 23 Apr 2012
- 43.Park S, Wong M, Marras SA, Cross EW, Kiehn TE, Chaturvedi V, Tyagi S, Perlin DS. Rapid identification of Candida dubliniensis using a species-specific molecular beacon. J Clin Microbiol. 2000;38:2829–36. doi: 10.1128/jcm.38.8.2829-2836.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Castelli MV, Buitrago MJ, Bernal-Martinez L, Gomez-Lopez A, Rodriguez-Tudela JL, Cuenca-Estrella M. Development and validation of a quantitative PCR assay for diagnosis of scedosporiosis. J Clin Microbiol. 2008;46:3412–6. doi: 10.1128/JCM.00046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Sanjay Tyaji 23 Apr 2012
- 45.Morozumi M, Nakayama E, Iwata S, Aoki Y, Hasegawa K, Kobayashi R, Chiba N, Tajima T, Ubukata K. Simultaneous detection of pathogens in clinical samples from patients with community-acquired pneumonia by real-time PCR with pathogen-specific molecular beacon probes. J Clin Microbiol. 2006;44:1440–6. doi: 10.1128/JCM.44.4.1440-1446.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Sanjay Tyaji 23 Apr 2012
- 46.Loens K, Beck T, Goossens H, Ursi D, Overdijk M, Sillekens P, Ieven M. Development of conventional and real-time nucleic acid sequence-based amplification assays for detection of Chlamydophila pneumoniae in respiratory specimens. J Clin Microbiol. 2006;44:1241–4. doi: 10.1128/JCM.44.4.1241-1244.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Sanjay Tyaji 23 Apr 2012
- 47.Chiba N, Murayama SY, Morozumi M, Nakayama E, Okada T, Iwata S, Sunakawa K, Ubukata K. Rapid detection of eight causative pathogens for the diagnosis of bacterial meningitis by real-time PCR. J Infect Chemother. 2009;15:92–8. doi: 10.1007/s10156-009-0670-3. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Sanjay Tyaji 23 Apr 2012
- 48.Saidac DS, Marras SA, Parveen N. Detection and quantification of Lyme spirochetes using sensitive and specific molecular beacon probes. BMC Microbiol. 2009;9:43. doi: 10.1186/1471-2180-9-43. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Sanjay Tyaji 23 Apr 2012
- 49.Elsayed S, Plewes K, Church D, Chow B, Zhang K. Use of molecular beacon probes for real-time PCR detection of Plasmodium falciparum and other plasmodium species in peripheral blood specimens. J Clin Microbiol. 2006;44:622–4. doi: 10.1128/JCM.44.2.622-624.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schneider P, Wolters L, Schoone G, Schallig H, Sillekens P, Hermsen R, Sauerwein R. Real-time nucleic acid sequence-based amplification is more convenient than real-time PCR for quantification of Plasmodium falciparum. J Clin Microbiol. 2005;43:402–5. doi: 10.1128/JCM.43.1.402-405.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Sanjay Tyaji 23 Apr 2012
- 51.Liming SH, Bhagwat AA. Application of a molecular beacon-real-time PCR technology to detect Salmonella species contaminating fruits and vegetables. Int J Food Microbiol. 2004;95:177–87. doi: 10.1016/j.ijfoodmicro.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 52.Heijnen L, Medema G. Method for rapid detection of viable Escherichia coli in water using real-time NASBA. Water Res. 2009;43:3124–32. doi: 10.1016/j.watres.2009.04.025. [DOI] [PubMed] [Google Scholar]
- 53.Nadal A, Coll A, Cook N, Pla M. A molecular beacon-based real time NASBA assay for detection of Listeria monocytogenes in food products: role of target mRNA secondary structure on NASBA design. J Microbiol Methods. 2007;68:623–32. doi: 10.1016/j.mimet.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 54.Huang Q, Hu Q, Li Q. Identification of 8 foodborne pathogens by multicolor combinational probe coding technology in a single real-time PCR. Clin Chem. 2007;53:1741–8. doi: 10.1373/clinchem.2007.087502. [DOI] [PubMed] [Google Scholar]
- 55.Hadjinicolaou AV, Demetriou VL, Emmanuel MA, Kakoyiannis CK, Kostrikis LG. Molecular beacon-based real-time PCR detection of primary isolates of Salmonella Typhimurium and Salmonella Enteritidis in environmental and clinical samples. BMC Microbiol. 2009;9:97. doi: 10.1186/1471-2180-9-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Singh G, Vajpayee P, Ram S, Shanker R. Environmental reservoirs for enterotoxigenic Escherichia coli in south Asian Gangetic riverine system. Environ Sci Technol. 2010;44:6475–80. doi: 10.1021/es1004208. [DOI] [PubMed] [Google Scholar]
- 57.Gatcombe RR, Jothikumar N, Dangoudoubiyam S, Kazacos KR, Hill VR. Evaluation of a molecular beacon real-time PCR assay for detection of Baylisascaris procyonis in different soil types and water samples. Parasitol Res. 2010;106:499–504. doi: 10.1007/s00436-009-1692-6. [DOI] [PubMed] [Google Scholar]
- 58.Gubala AJ, Proll DF. Molecular-beacon multiplex real-time PCR assay for detection of Vibrio cholerae. Appl Environ Microbiol. 2006;72:6424–8. doi: 10.1128/AEM.02597-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fykse EM, Skogan G, Davies W, Olsen JS, Blatny JM. Detection of Vibrio cholerae by real-time nucleic acid sequence-based amplification. Appl Environ Microbiol. 2007;73:1457–66. doi: 10.1128/AEM.01635-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hadjinicolaou AV, Demetriou VL, Hezka J, Beyer W, Hadfield TL, Kostrikis LG. Use of molecular beacons and multi-allelic real-time PCR for detection of and discrimination between virulent Bacillus anthracis and other Bacillus isolates. J Microbiol Methods. 2009;78:45–53. doi: 10.1016/j.mimet.2009.04.005. [DOI] [PubMed] [Google Scholar]