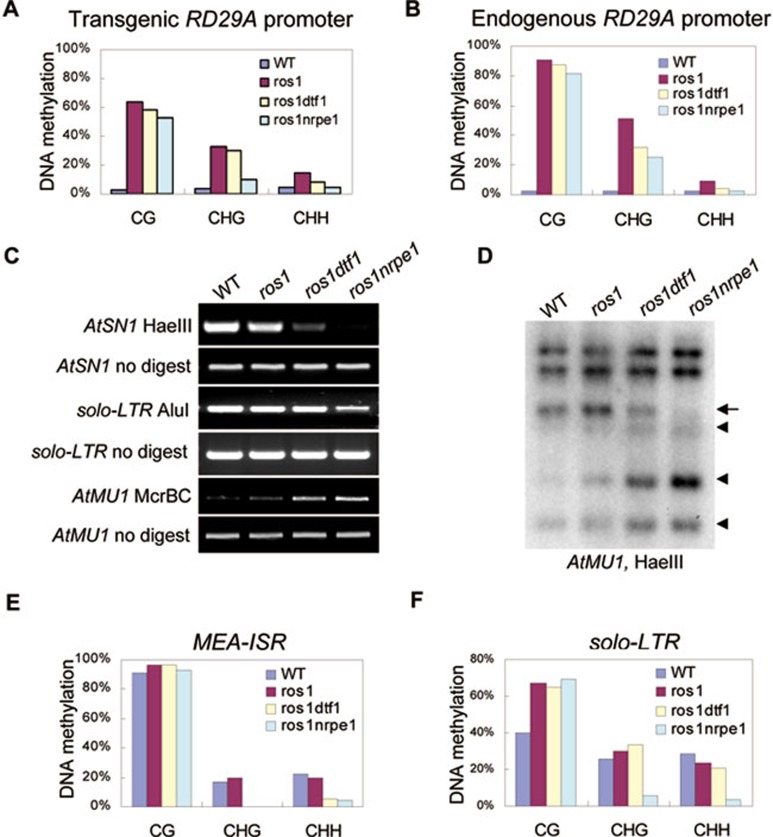
Figure 4.
Effect of dtf1 on DNA methylation. (A, B) The effect of dtf1 on the promoter DNA methylation of RD29A-LUC (A) and endogenous RD29A (B) was determined by bisulfite sequencing. The percentage of methylated cytosine in different cytosine contexts is shown. (C) The DNA methylation level of AtSN1, solo-LTR and AtMU1 was determined by chop-PCR. The DNA methylation-sensitive restriction enzymes HaeIII and AluI, and DNA mehylation-insensitive enzyme McrBC were used for DNA methylation analysis as indicated. (D) The effect of dtf1 on AtMU1 DNA methylation was detected by Southern blot analysis. Genomic DNA from the indicated genotypes was digested by HaeIII followed by Southern hybridization. (E and F) The DNA methylation status of MEA-ISR and solo-LTR was determined by bisulfite sequencing in the indicated genotypes.