Figure 2.
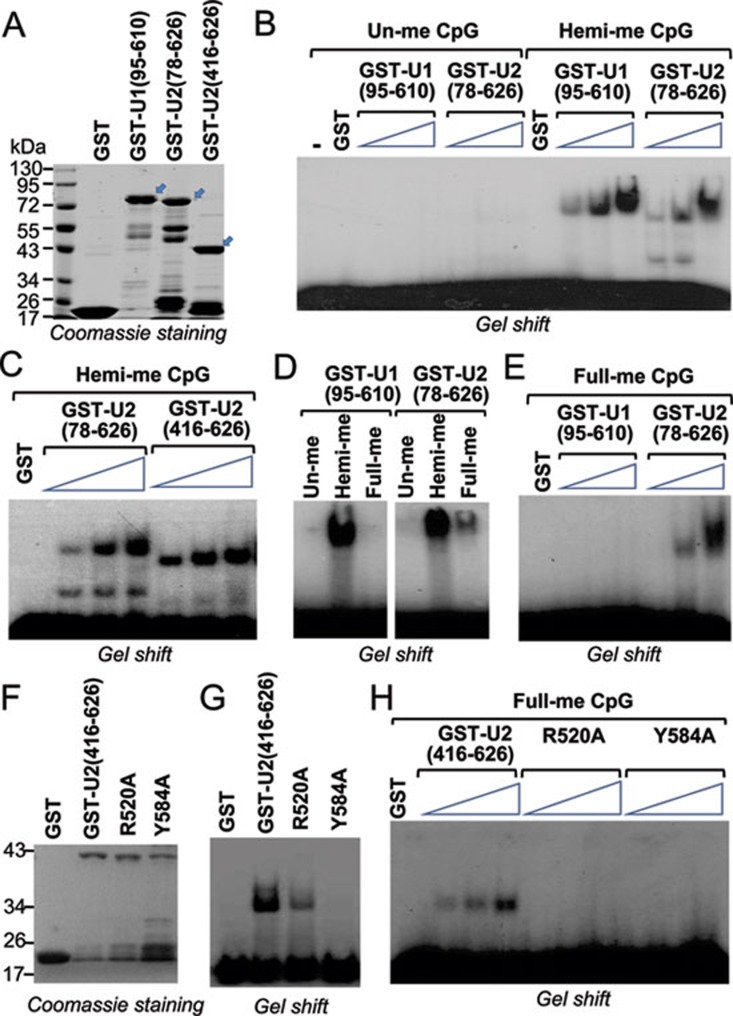
UHRF2 binds hemi- and fully methylated DNA through its SRA domain. (A) Coomassie blue staining gel showing purified GST-fusions of UHRF1 and UHRF2. The regions of UHRF1 or UHRF2 in number of amino acids fused to GST were as indicated. Arrows indicate the positions of GST-fusion proteins with the expected sizes. Also indicated are protein size markers. (B) Gel mobility shift assay showing binding of GST-UHRF1 (95-610) and GST-UHRF2 (78-626) to hemi-methylated but not un-methylated DNA probes. An increasing amount of recombinant proteins (0.25 μg, 0.5 μg and 1 μg) were used for gel shift. (C) The recombinant UHRF2 containing SRA domain alone, GST-UHRF2 (416-626), is capable of binding hemi-methylated DNA probe. 0.25, 0.5 and 1 μg of proteins were used for gel shift. (D) Comparison of GST-UHRF1 (95-610) and GST-UHRF2 (78-626) in binding of un-methylated, hemi-methylated (one strand methylated) and fully methylated (both strands methylated) DNA probe. (E) Increasing amount of GST-UHRF1 (95-610) and GST-UHRF2 (78-626) were compared for binding of fully methylated DNA probe. Three concentrations used were 0.25, 0.5 and 1 μg, respectively. (F) Coomassie blue staining gel showing purified GST, GST-UHRF2 (416-626), and GST-UHRF2 (416-626) with R520A or Y584A mutation. (G) Gel mobility shift assay showing impaired hemi-methylated DNA binding activity for R520A and Y584A mutants. (H) R520A and Y584A mutants also showed impaired binding activity for fully methylated DNA probe. Three concentrations of proteins used were 0.25, 0.5 and 1 μg, respectively.