Abstract
Zebrafish (Danio rerio) are an increasingly important biological model in many areas of research. Diseases of zebrafish, especially those resulting in chronic, sub lethal infections, are of great concern due to the potential for non-protocol induced variation. The microsporidium, Pseudoloma neurophilia, is a common parasite of laboratory zebrafish. Current methods for detection of this parasite require lethal sampling of fish, which is often undesirable with poorly spawning mutant lines and small populations. We present here an improved molecular based diagnostic assay using real-time PCR, and including sonication treatment prior to DNA extraction. Sensitivity was increased compared to the previously published conventional PCR-based assay based on a dilution experiment, showing that this new assay had the ability to detect parasite DNA in one log higher dilution than the conventional PCR-based assay, which did not include sonication. Comparisons of several DNA extraction methods were also performed to determine the method providing the maximum sensitivity. Sonication was found to be the most effective method for disrupting spores. Further, we demonstrate the application of this method for testing of water, eggs and sperm, providing a potential non-lethal method for detection of this parasite in zebrafish colonies with a sensitivity of 10 spores per liter, 2 spores per egg and 10 spores per μl of sperm, respectively.
Keywords: microsporidia, real-time PCR, Danio
INTRODUCTION
The zebrafish, Danio rerio, is a widely used biological model in several fields including developmental biology, immunology, toxicology, infectious disease and cancer research (Eisen 1996, Dooley & Zon 2000, Amatruda et al. 2002, Hill et al. 2005). Laboratory colonies of zebrafish are typically composed of specialized mutant strains of fish possessing genotypes useful to specific areas of study, and hardier wild-type strains are used for breeding stock and for maintaining specific genotypes in a heterozygotic state (Westerfield 2007). Maintenance and husbandry of many of the mutant strains is often difficult (Lawrence 2007). Individual adult fish from these lines, therefore, are often in limited supply and may be extremely valuable. Whereas laboratory populations of zebrafish occasionally are affected by acute infectious diseases, the most important and more prevalent are chronic infections by Mycobacterium spp. and Pseudoloma neurophilia (Microsporidia) (Matthews 2004, Kent et al. 2009). The latter has been detected in over 50% of the facilities that we have examined through the Zebrafish International Resource Center (ZIRC) diagnostic service.
Pseudoloma neurophilia, a microsporidian parasite, generally causes chronic infections in zebrafish with clinical signs ranging from obvious scoliotic changes and emaciated appearance of fish to subclinical infections exhibiting no outward signs of disease (Matthews et al. 2001). As with other animals used in research, experiments utilizing zebrafish with these infections are subject to non-experimental variation, potentially confounding results as described in laboratory colonies of rabbits and mice infected with the microsporidian parasite, Encephalitozoon cuniculi (Baker 1998). Furthermore, fish without overt clinical disease may have reduced fecundity associated with the infection (Ramsay et al. 2009).
Microsporidia are obligate intracellular parasites with species infecting virtually all animal phyla. They have a relatively simple life cycle, consisting of two general developmental stages; mergony and sporogony. Meronts multiply inside the infected host cell, eventually forming sporonts and then spores, which are ultimately released from the host and transmit the infection. The infectious spore stage has a thick, chitinous outer layer, making it extremely resistant to environmental stress and lysis, allowing the organism to maintain viability for extended periods in the aquatic environment (Shaw et al 2000).
While several assays exist for the detection of pathogens in other fish species using non-lethal sampling methods (Miriam et al. 1997, López-Vázquez et al. 2006, Lindstrom et al. 2009), these sampling methods are generally not applicable to the zebrafish due to its small size and difficulty in obtaining blood and other tissues. Because zebrafish can be housed in relatively small volume tanks, the screening of water in tanks and even effluent from flow-through systems seems a feasible method by which to detect this pathogen without requiring lethal sampling of adult fish. Also, zebrafish spawn frequently and thus sperm, eggs, etc. are readily available providing potential samples for testing.
For Microsporidia infecting fish, the only viable stage found in water is the spore, as other stages of these obligate intracellular parasites would not survive long outside the host. Therefore, in contrast to presporogonic stages found in tissues, efficient disruption of spores is crucial for obtaining DNA in the development of sensitive assays that are based on the detection of this stage. Disruption of the tough, chitinous spore stage of the parasite requires special methods such as mechanical disruption by bead beating or sonication (Docker et al. 1997, Dowd et al. 1998, Müller et al. 1999, Fournier et al. 2000, Graczyk et al. 2007, Hoffman et al. 2007, Phelps & Goodwin 2007), the use of saccharolytic enzymes such as chitinase (Müller et al. 1999, Delarbre et al. 2001), or in vitro germination of the spore using chemical means such as hydrogen peroxide in combination with other purification methods (Higes et al 2006). Additionally, the few genomic studies of microsporidian parasites have found a general pattern of small, streamlined genomes with very few gene copies (Williams et al. 2008). In fact, one genus has been found to possess a single copy of the small subunit ribosomal RNA gene (Cornman et al. 2009), highlighting the need to maximize spore disruption and DNA concentration to achieve a sensitive and practical method of detection as most PCR tests for microsporidia have been based on this gene (Zhu et al. 1993, Brown & Kent 2002, Joseph et al. 2006).
Whipps and Kent (2006) developed a PCR test for P. neurophilia based on the detection of small subunit ribosomal RNA (ssrDNA) gene sequences, and it was capable of consistently detecting down to an estimated 10 spores/sample of brain and spinal cord tissue. Here we describe a new more sensitive PCR test for P. neurophilia, using a real-time PCR platform. Our intent was to use this assay for screening water and sex products and an important focus of this study was determination of optimal extraction methods for detecting the spores in both media. We also included screening of water from spawning tanks, eggs and sperm for non-lethal testing as zebrafish spawn prodigiously and spores of P. neurophilia occur both within and outside eggs in ovaries (Kent & Bishop-Stewart 2003, Kent et al. 2007).
MATERIALS AND METHODS
Pseudoloma neurophilia spore preparations
All Pseudoloma neurophilia spores used in this study were obtained from hind brain and anterior spinal cords from 40 known infected stock zebrafish that were euthanized by an overdose of tricaine methanesulfite (Argent Laboratories, Redmond, WA). A modification of a previously described method (Ferguson et al. 2007) was used to isolate spores. Briefly, hindbrain and spinal cord tissue were mixed with 5 ml sterile phosphate buffered saline (PBS) and then homogenized by passing through successively smaller gauge needles and filtered through a 20 μm nylon mesh filter. This homogenate was then centrifuged through a 50% Percoll (Sigma-Aldrich, St. Louis, MO) gradient for 50 minutes at 1,200 g and further purified by washing in a 0.45 μm filter (Millipore, Billerica, MA) twice with (PBS) to minimize the number of presporogonic stages present, in order for results of the extraction studies to be based on spores only. The resulting spore suspension was eluted from the filter in PBS, quantified using a hemocytometer and then diluted in PBS as needed.
Assay design
A Taqman-based PCR assay was used to measure P. neurophilia DNA using an ABI 7500 sequence detection system (ABI Biosystems, Foster City, CA). Primers were designed to amplify the ssuRNA gene of P. neurophilia based on sequence available in the NCBI Genbank (accession number AF322654) and using the Primer-BLAST program also available online from the NCBI with the primer parameters set to search for a PCR product size from 70-125 bp, optimal primer melting temperature of 60°C, and BLAST parameters set to search for similarity in the NCBI non-redundant database (All GenBank + RefSeq Nucleotides + EMBL + DDBJ + PDB sequences (excluding HTGS0,1,2, EST, GSS, STS, PAT, WGS) ) for specificity. The forward primer Pn10F (5’ GTAATCGCGGGCTCACTAAG 3’), and reverse primer Pn10R (5’ GCTCGCTCAGCCAAATAAAC 3’) were selected on the basis of lack of identity to related microsporidia, annealing temperature and small amplicon size of 113 base pairs, from position 1175 to position 1288 on the ssuRNA gene. A 3’ hydrolysis probe complementary to a 23 base pair section of the amplified region was designed using the sequence 5’- 6-carboxyfluorescein (FAM)-ACACACCGCCCGTCGTTATCGAA – 3’-Black Hole Quencher 1 (BHQ1). All reactions were performed in 25 μl using 900 nM forward and reverse primers, 250 nM of hydrolysis probe, 1X TaqMan Universal PCR Master Mix (ABI) and 2 μl of sample extract. The real-time PCR was performed on an ABI 7500 Sequence Detection system (ABI) using the following reaction conditions: 50°C for 2 minutes, 95°C for 10 minutes followed by 40 repetitions (95°C for 15s, 60°C for 1 minute). Data analysis was performed using the 7500 System SDS Software version 1.3.1 (ABI).
Cross reactivity testing
Cross reactivity of the assay was performed using the PN10F/PN10R primer and the PN probe with DNA extracts obtained from spores of two fish microsporidian parasites that could potentially be encountered in a zebrafish research facility: Glugea anomala obtained from three-spined sticklebacks in a research colony and Pleistophora hyphessobryconis obtained from neon tetras from a commercial tropical fish vendor.
Evaluation of pretreatments and DNA extraction methods for spores
Sonication
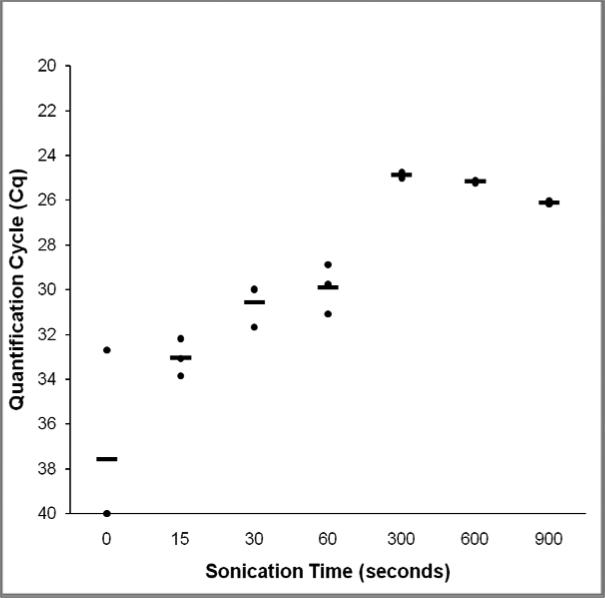
Sonication was one of our primary pretreatment methods to test. Prior to comparing pretreatment and extraction methods in the two trials below, it was necessary to determine the optimal sonication time for the disruption of spores. Thus a time study was performed in which several suspensions of spores consisting of 1,000 spores in 100 μl of PBS were sonicated with a Branson Sonifier 250 (Branson, Danbury, CT) for various time points and 2 μl of the crude sonicate tested by the real-time PCR method described above. The time points tested were 15 – 900 sec (Figure 2) and all were run at 55 W at a frequency of 20 kHz. Samples were not cooled during the sonication period, but were immediately placed on ice after treatment. The probe was decontaminated between samples using 10% household bleach followed by rinsing with sterile water. Each time point was examined in triplicate. It was determined that 5 min consistently provided the lowest quantification cycle and thus this was used for the sonication method in the extraction comparison trials.
Fig. 2.
Pseudoloma neurophilia spore sonication time study. Quantification cycle thresholds (Cq) of triplicate suspensions of 1000 spores of P. neurophilia sonicated at 55W for different time points and tested by real-time PCR. Bars represent mean Cq; Points represent individual sample Cqs.
Trial A
A preliminary experiment was performed to assess the efficacy of sonication and hydrogen peroxide pretreatments in conjunction with three DNA extraction methods (Table 1). Trial A was performed using an initial 900 μl suspension containing 30,000 spores in total. The suspension was divided into three, 300 μl aliquots and subjected to either no treatment or one of two pretreatments:
Table 1.
Experimental setup for two extraction comparison trials. Trial A samples represent 300 μl aliquots taken from an initial pool of 30,000 spores in 900 μl of PBS. Following pretreatment, samples were divided into 6 aliquots for DNA extraction. Each extraction method was performed twice. Samples in Trial B represent individual purified spore suspensions containing 1,000 spores each in 100 μl PBS. Methods in Trial B were performed in triplicate.
| Trial | Sample | Pretreatment | DNA Extraction |
|---|---|---|---|
| A | 1 | NT | No Extraction |
| QIAgen | |||
| MoBio | |||
| 2 | Hydrogen Peroxide | No Extraction | |
| QIAgen | |||
| MoBio | |||
| 3 | Sonication | No Extraction | |
| QIAgen | |||
| MoBio | |||
| B | 1 | None | None |
| 2 | Bead Beating | QIAgen | |
| 3 | Chitinase | QIAgen | |
| 4 | None | QIAgen | |
| 5 | None | MoBio | |
| 6 | Sonication | None | |
| 7 | Sonication | QIAgen | |
Pretreatment with hydrogen peroxide
As numerous microsporidian spores have been shown to germinate in vitro in the presence of hydrogen peroxide (Keohane & Weiss 1999) we attempted to germinate spores of P. neurophilia by adding 30 μl of 30% hydrogen peroxide to the spore sample for a final concentration of 9% and incubated at 30°C for 30 minutes. The suspension was centrifuged for 5 minutes at 14,000 × g in a tabletop centrifuge. The supernatant was removed, 300 μl of molecular grade water was added and the suspension was allowed to sit for 30 minutes prior to further treatment.
Pretreatment with sonication
Samples were sonicated with a Branson Sonifier 250 (Branson, Danbury, CT) for 5 minutes and immediately placed on ice after treatment. The probe was decontaminated between samples using 10% household bleach followed by rinsing with sterile water.
Following pretreatment (none, sonication or hydrogen peroxide), each pretreated pool was then divided into six 50 μl aliquots and either tested directly or extracted in duplicate using the following methods before testing duplicate reactions by real-time PCR:
QIAgen
The DNeasy Blood and Tissue Extraction kit (QIAgen, Inc., Valencia, CA) was used for this procedure following the manufacturer's protocol for extraction of DNA from tissues, with the addition of a single overnight freeze-thaw of the spore suspension and an overnight proteinase K and lysis buffer digestion at 56°C. Samples were eluted in 100 μl of buffer supplied in the kit.
MoBio
The UltraClean™ Microbial DNA Isolation kit (Mo Bio, Carlsbad, CA) employs heat, detergent lysis, and bead-beating using specialized bead tubes and a standard vortex mixer with an adapter plate. The manufacturer's protocol was followed using the included reagents, bead tubes, and silica membrane centrifugal filter columns. Briefly, 50 μl of pretreated/nonpretreated spore suspensions were first added to detergent buffer and heated at 65°C for ten minutes after which they were placed in the bead tubes and vortexed for another ten minutes. After vortexing, the DNA was bound to a silica membrane centrifugation filter and cellular components were washed out with provided wash buffer. Finally, DNA was eluted in 50 μl of tris buffer supplied in the kit.
Trial B
Based on the results from Trial A, pretreatment with hydrogen peroxide was not used and sonication was investigated further in a second experiment which was performed using purified spore suspensions containing 1,000 spores each in 100 μl PBS. Here, pretreatment with bead beating and chitinase was tested, along with sonication (as described in Trial A) and no pretreatment (Table 1). QIAgen (with or without pretrements) and MoBio extractions (according manufacturer instructions) were conducted as described in Trial A. Three spore suspensions were tested directly by real-time PCR with no pretreatment nor DNA extraction and the following extraction methods were compared using triplicate spore suspensions:
Bead beating
Spores were suspended in Buffer ATL with proteinase K, both from the QIAgen Blood and Tissue Extraction kit, and 500 mg of 0.5 μm glass beads (Sigma-Aldrich, St. Louis, MO) in 1.5 ml screw-cap tubes. The samples were then run on a bead beater (BioSpec, Bartlesville, OK) at high speed (4500 oscillations per minute) for 3 minutes after which they were incubated for two hours at 56°C and extracted following the QIAgen method described above. All samples were eluted in a final volume of 100 μl of buffer supplied in the QIAgen kit.
Chitinase
Chitinase (0.4U, Sigma-Aldrich, St. Louis, MO) and 200 μl potassium phosphate buffer (200 mM, pH 6.0) was added to each sample which was then incubated at 37°C for 30 minutes. Proteinase K from the Dneasy kit was then added and the samples were processed using the QIAgen method per manufacturer's protocol and eluted in a final volume of 100 μl of buffer supplied in the QIAgen kit.
Statistical analysis
Levene's test for equality of variances was performed on the Cq values obtained in Trial B. After determining that there were no significant differences between variances in the extraction methods (Levene's test, p=0.73), a One-way Analysis of Variance (ANOVA) was performed. Multiple comparison with best procedure based on Hsu's method (Kuehl 2000) was performed to determine the methods which provided the highest sensitivity. All analyses were performed using the statistical package R (www.r-project.org).
Based on results from the extraction method study, all further experiments employed sonication or sonication followed by QIAgen extraction.
Detection limit of real-time PCR
Spores in PBS
A spore suspension was prepared as described above and spore suspensions were made in triplicate using 1,000 spores in 100 μl PBS, 100 spores/ 100 μl PBS, and 10 spores/ 100 μl PBS. Each suspension was sonicated and 2 μl of the crude sonicate was directly analyzed using the real-time PCR method described above in order to obtain 20, 2, and 0.2 spores per real-time PCR reaction, respectively.
Spores in Group Spawn Tank Water
The ability of the test to detect the parasite in spawn water was evaluated. Ten liters of fish system water was divided into 1 l flasks and inoculated as follows: three 1 l aliquots with 10 spores, three 1 l aliquots with 50 spores, three 1 l aliquots with 100 spores, three 1 l aliquots with 500 spores and one 1 l aliquot remained as a negative control.
Each spiked 1 l water sample was individually filtered through a 1.2 μm nitrocellulose filter (Millipore #RAWP04700) in a fritted glass filter holder (Millipore, Billerica, MA) using a vacuum pump at 300 mm Hg. After filtration, the filter was rolled up using sterile forceps and placed in a 1.5 ml conical tube screw-cap tube. The water filtration apparatus was washed and decontaminated with 10% household bleach followed by rinsing in sterile water between samples.
One ml of acetone was added to each 1.5 ml conical tube with the nitrocellulose filter and vortexed for several seconds. The tubes were then centrifuged at maximum speed for 3 minutes (> 16,000 rcf). The acetone supernatant was carefully aspirated off of the pellet with a transfer pipette. This was repeated two times to ensure all dissolved nitrocellulose was removed from the sample. One ml of 100% ethanol was then added to each tube and the pellet was suspended by vortexing and centrifuged at 3,000 g for 5 minutes. The ethanol was again carefully aspirated and 5 ml of 70% ethanol was added to each tube. Tubes were centrifuged for 5 minutes at 3,000 g and the ethanol was again aspirated. The pellet was resuspended in 100 μl of phosphate buffered saline (PBS) and then was sonicated for 5 minutes at 55W. After sonication, each sample was placed on ice and DNA was extracted using the QIAgen extraction protocol described above. All samples were eluted in a final volume of 200 μl of buffer supplied in the kit.
Spores in Eggs
We conducted the following test to evaluate detection of spores associated with eggs. A total of 2,000 eggs were obtained from the SPF zebrafish colony at the Sinnhuber Aquatic Research Laboratory (SARL), Oregon State University and divided into four aliquots of 500 eggs each with PBS added to make a total volume of 1 ml. One aliquot was spiked with 100 spores, one with 1000 spores, one with 10,000 spores and the last aliquot had no spores (negative control). Each sample was then sonicated for 5 minutes at 55W and cooled on ice. After sonication, three 40 μl aliquots (equaling approximately 20 eggs based on the original volume) were taken from each sample and DNA was extracted using the QIAgen blood and tissue protocol as described above. All samples were eluted in a final volume of 200 μl of buffer supplied in the kit.
Spores in Sperm
Sperm was obtained from SPF zebrafish males from the colony at the SARL by squeezing to evaluate the test with this sex product. The sperm samples were pooled and divided into 5 μl aliquots in 100 μl of PBS and spiked, in triplicate, with 5 spores/sample, 50 spores/sample and 500 spores/sample. All samples were brought to a volume of 200 μl and were then sonicated and DNA was extracted using the QIAgen extraction described above. All samples were eluted in a final volume of 200 μl of buffer supplied in the kit. The spiked spawn water, eggs and sperm were run in triplicate on different days.
Comparison with conventional PCR assay
We compared our new real-time PCR based assay, which includes sonication pretreatment, with the previously developed conventional PCR based test described by Whipps and Kent (2006), which does not include sonication. The hindbrain and spinal cords from six infected fish were removed and individually homogenized in 100 μl of sterile PBS. Then 50 μl of the resulting homogenate was sonicated for 5 minutes followed by purification using the QIAgen method and the remaining 50 μl was extracted by the QIAgen method as described by Whipps and Kent (2006). All DNA extracts were eluted in 100 μl of tris buffer and four serial tenfold dilutions of each was made in sterile water. After extraction, all samples and dilutions were run in single reactions using real-time PCR as described here and the conventional method as described by Whipps and Kent (2006) with minor modification. Briefly, the primer pair Pn18S5F 5′ GAA AAT TAC CGG AGC CTG AAG TC 3′, and Pn18S5R, 5′ TTC CCT CTC TCT CCA AAT TTC GG 3′ were used to amplify a 788 bp fragment of the ssrDNA of P. neurophilia using conventional PCR. The reaction was carried out in 25 μL volumes using the Platinum® PCR SuperMix (Invitrogen, Carlsbad, CA), 12.5 pmol of each primer and 2 μl of extracted DNA. Amplification was carried out on a PTC-200 thermocycler (MJ Research, Watertown, MA) with an initial denaturation at 94°C for 3 minutes followed by 35 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 60s, and a final extension step of 72°C for 7 minutes. Products were visualized on a 1.5% agarose gel stained with SYBR® Safe DNA Gel Stain (Invitrogen, Carlsbad, CA). Results were reported as positive (P. neurophilia ssrDNA detected) or negative (no P. neurophilia ssrDNA detected) based on the presence or absence of a band corresponding with an approximate size of 788 base pairs.
Group Spawn Experiment
The ability of the test to detect the parasite in sex products of infected fish was evaluated. Ten adult zebrafish were arbitrarily selected from a population determined to have a 10% prevalence of Pseudoloma neurophilia based on histological examination of a subsample of fish three months prior to the experiment. The fish were placed in a spawning tank with 10 l of system water overnight. The following day, the fish were collected and euthanized by an overdose of tricaine methanesulfonate (Argent Laboratories, Redmond, WA). Brains and spinal tissue were collected using sterile instruments between individuals, placed in separate 1.5 ml tubes and extracted using the standard QIAgen protocol with an overnight digestion at 56°C. Water was filtered in 1 l aliquots using the method described previously. Filters were dissolved, sonicated and extracted as described previously. Eggs were pooled, sonicated and five 40μl aliquots were extracted as described previously. All samples were tested in single reactions using the real-time PCR method.
RESULTS
Cross Reactivity
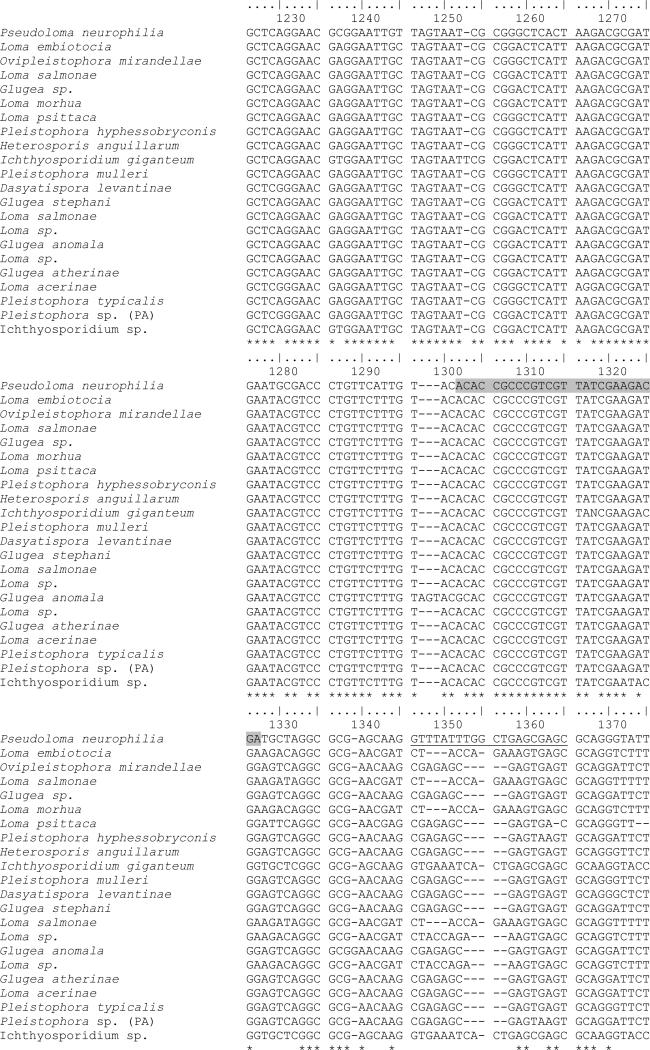
Primer-BLAST search of the 20 base pair forward primer, PN10F and the 20 base pair reverse primer, PN10R returned AF322654.1 Pseudoloma neurophilia small subunit ribosomal RNA gene, partial sequence; internal transcribed spacer, complete sequence; and large subunit ribosomal RNA gene, partial sequence, with no other matches found in the selected database (All GenBank+EMBL+DDBJ+PDB sequences but no EST, STS, GSS, environmental samples or phase 0, 1 or 2 HTGS sequences). An alignment of partial ssrDNA sequences of several related Microsporidia showed several nucleotide mismatches to that of P. neurophilia present within the 113 base pair amplicon in regions specific to both primers and probe sequences (Figure 1). Testing of DNA extracted from spores of Glugea anomala and Pleistophora hyphessobryconis resulted in no increase in fluorescence detected through 40 cycles of real-time PCR.
Fig. 1.
Partial ssrDNA sequence alignment of Pseudloma neurophilia (Genbank AF322654), Loma embiotocia (Genbank AF320310) , Ovipleistophora mirandellae (Genbank AF356223), Loma salmonae (Genbank U78736), Glugea sp. (Genbank AY090038), Loma morhua (Genbank GQ121037), Loma psittaca (Genbank FJ843104), Pleistophora hyphessobryconis (Genbank GU126672), Heterosporis anguillarum (Genbank AF387331), Ichthyosporidium giganteum (Genbank L13430), Pleistophora mulleri (Genbank FN434084), Dasyatispora levantinae (Genbank GU183263), Glugea stephani (Genbank AF056015), Loma salmonae (Genbank HM626215), Loma sp. (Genbank HM626217), Glugea anomala (Genbank AF044391), Loma sp.(Genbank AF104081), Glugea atherinae (Genbank U15987), Loma acerinae (Genbank AJ252951), Pleistophora typicalis (Genbank AJ252956), Pleistophora sp (PA) (Genbank AJ252958), and Ichthyosporidium sp.(Ganbank L39110) Pn10F and Pn10R primer locations are underlined and PN10 probe location is shaded. Asterisks denote regions of nucleotide similarity.
Sonication Time Study
Sonication for 5 min at 55W was determined to be the optimal lysis time to ensure consistently high extraction efficiency of Pseudoloma neurophilia spores with minimal loss of signal due to DNA shearing (Figure 2).
Extraction Comparison
In Trial A, the sonication pretreatment, both unextracted (mean Cq = 26.9) and followed by QIAgen extraction (mean Cq = 27.7), resulted in a lower mean crossing threshold than other methods (Figure 3). The multiple comparison with best procedure analysis of Trial B showed chitinase pretreatment followed by QIAgen extraction (CQ), QIAgen extraction with no pretreatment (Q), sonication pretreatment with no further extraction (S) and sonication pretreatment followed by QIAgen extraction (SQ) to be in the best group with sonication alone (S) to have the lowest mean crossing threshold (mean Cq = 28.74). Sonication followed by QIAgen extraction (mean Cq = 29.37) had the next lowest mean crossing threshold (Figure 3).
Fig. 3.
Pseudoloma neurophilia spore extraction method comparison by quantification cycle (Cq). NT: no treatment, M: MoBio, Q: QIAgen, P: peroxide, P+M: peroxide + Mobio, P+Q: peroxide + QIAgen, S: sonication, S+M: sonication + Mobio, S+Q: sonication + QIAgen, B+Q: bead beating + QIAgen, C+Q: chitinase + QIAgen. Bars represent mean Cq; points represent actual Cq for individual samples. Sonication treatment alone showed the lowest mean Cq in Trial A, 26.74 followed by sonication with DNA extraction by QIAgen silica gel membrane method (mean Cq = 27.73). Trial B results showed the lowest mean Cq for sonication alone, 28.74, followed by sonication and QIAgen DNA extraction combined, 29.93.
Detection Limit
The assay, using the PN10F/R primer set and probe PN, consistently detected less than one spore (0.2 spore) per reaction in PBS when the sample was sonicated. This was determined based on the original number of spores (10 spores/100 μl), 2 μl of which were tested by real-time PCR. Pseudoloma neurophilia ssrDNA was detected when spawn water, eggs or sperm samples were spiked with spores at 4-5 spores per real-time PCR reaction in all trials and replicates within trials (Table 2) using sonication followed by Qiagen extraction. Parasite ssrDNA was detected in spawn water spiked with as low as 10 spores per liter (0.1 spore per reaction) in one trial, however, it was not detected in the other two. We detected the parasite in eggs spiked with 2 spores per egg (0.4 spore per reaction) in all trials and in all but one replicate. We also detected the parasite in spiked sperm samples, consistently detecting the parasite at 10 spores per μl of sperm (0.5 spore per reaction). The inconsistent detection at these lower levels of parasite is likely reflective of sampling error inherent in dealing with such dilute concentrations.
Table 2.
Pseudoloma neurophilia spiked sample results from three separate trials. Numbers for each trial represent quantification cycle threshold (Cq) detected by real-time PCR per sample. Spawn water consists of purified spores of P. neurophilia in 1 l of water from a group spawn of P. neurophilia free (SPF) zebrafish. Eggs consist of purified spores of P. neurophilia in a 40 μl aliquot (representing approximately 20 eggs based on an initial volume of 500 eggs/lml) of homogenate made from eggs spawned by SPF zebrafish. Sperm samples consist of purified spores of P. neurophilia in a 5 μl aliquot of sperm obtained by squeezing SPF zebrafish males. - : no copies of P. neurophilia ssrDNA detected after 40 cycles.
| Sample Type | Starting Concentration (Spores per reaction) | Trial 1 | Trial 2 | Trial 3 | Mean (Standard Deviation) |
|---|---|---|---|---|---|
| Spawn Water | 500 (5) | 36.7, 35.0, 36.4 | 35.8, 34.8, 37.9 | 35.6, 36.1, 37.9 | 36.2(1.1) |
| 100 (1) | -,-,39.9 | 39.4, 37.4, 38.8 | 38.8,39.3,39.7 | 39.0(0.8) | |
| 50 (0.5) | 38.3, 37.5, 39.4 | 38.9, -, - | 37.3, -, 39.4 | 38.5(0.9) | |
| 10 (0.1) | -,-,- | -,-,- | 38.1, 38.5, - | 38.3(0.3) | |
| Eggs | 400 (4) | 35.6, 36.3, 35.3 | 36.6, 37.9,36.7 | 36.1, 36.5,36.2 | 36.4(0.7) |
| 40 (0.4) | 38.6, -, - | 36.2, 36.8, 36.0 | 35.2, 36.9, 36.9 | 36.7(1.1) | |
| 4 (0.04) | -,-,- | 37.2, 38.6, 37.3 | 36.6, 37.6,- | 37.5(0.7) | |
| Sperm | 500 (5) | 32.7, 32.3, 31.9 | 35.5, -, 34.4 | 32.8,32.1, 31.3 | 32.9(1.4) |
| 50 (0.5) | 35.9, 36.6, 34.8 | 38.0, -, 34.4 | 35.6, 35.6, 34.9 | 35.7(1.2) | |
| 5 (0.05) | 36.4, -, 36.3 | 36.8, -, 34.4 | 35.1,-,- | 35.8(1.0) | |
Comparison to Conventional Assay
With the exception of one tissue sample, the real-time PCR assay detected P. neurophilia ssrDNA in at least one log higher dilution compared to the conventional assay (Table 3). Analysis of all six samples by the real-time PCR method showed that sonication followed by DNA extraction using the QIAgen DNeasy Blood and Tissue Extraction kit resulted in a mean decrease of crossing threshold of 2.34 compared to the same samples that were not sonicated.
Table 3.
Comparison of Pseudoloma neurophilia detection by new real-time PCR assay utilizing sonication pretreatment of samples with conventional PCR assay described by Whipps and Kent (2006). The real-time PCR assay described detected the parasite from one to three log dilutions higher than the conventional assay (+ = P. neurophilia ssrDNA detected, - = No P. neurophilia ssrDNA detected).
| Individual | Conventional PCR | Real-time PCR | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | -1 | -2 | -3 | -4 | 0 | -1 | -2 | -3 | -4 | |
| 1 | + | - | - | - | - | + | + | + | - | - |
| 2 | + | - | - | - | - | + | - | - | - | - |
| 3 | + | + | - | - | - | + | + | + | + | - |
| 4 | + | - | - | - | - | + | + | + | - | - |
| 5 | + | - | - | - | - | + | + | - | - | - |
| 6 | + | - | - | - | - | + | + | - | - | - |
Group Spawn Experiment
P. neurophilia ssrDNA was detected in the brain and spinal tissues of six of ten adult fish in the group spawning experiment. Eggs were pooled and divided into five aliquots and P. neurophilia ssrDNA was detected in one pool. In contrast, P. neurophilia ssrDNA was detected in nine of ten 1 l water samples from the spawning tank.
DISCUSSION
Our new real-time PCR test for Pseudoloma neurophilia provides a sensitive test for evaluation of fish tissues and a non-lethal method for detecting spores in fish tissues, water and in spawning products. This is best illustrated by the decreased crossing threshold (Cq) of samples using methods employing sonication (Figure 3), indicating an increase in the number of ssrDNA copies present. Comparing the test to the conventional PCR based assay developed by Whipps and Kent (2006), we found that it was 10-100 times more sensitive and our calculations indicated that it could detect < 1 spore per reaction based on the number of spores spiked into PBS. This increased sensitivity was not necessarily due to conventional versus real-time format or differences in primers, but more likely due to pretreatment by sonication in our new test as discussed below.
Regarding specificity, we designed the primers so that they would be unique to P. neurophilia. Also, we evaluated our test with Glugea anomala from 3-spined sticklebacks, Gasterosteus aculeatus, and Pleistophora hyphessobryconis from neon tetras, Paracheirodon innesi, as these two Microsporidia might be found in fish research facilities. Three-spined sticklebacks are used in laboratory research and we recently detected P. hyphessobryconis in three zebrafish colonies (Sanders et al. 2010). Additionally, a total of 278 fish from 6 separate populations housed in a known Pseudoloma neurophilia free zebrafish colony were tested using this real-time PCR based assay, with histological analysis on individuals from the same populations performed in parallel (see Kent et al. In Press). No fish from this population were positive for P. neurophilia by real-time PCR or histology.
Whereas the purpose of the present study was to develop a real-time PCR based assay, we found that the primer set used in this format also will work in a conventional PCR format as a positive/negative screening test. However, we did not determine the sensitivity of the primers using this conventional PCR format. Whereas quantification is not necessary for detection of the parasite, it may be pertinent to implement this feature for future studies on disease progression, transmission and dose response, thus the real-time PCR platform was used. Furthermore, the use of the real-time PCR platform eliminates post-amplification handling of samples, decreasing the chance of cross-contamination by amplicons leading to false positive results.
Current protocols for the detection of Pseudoloma neurophilia involve either direct observation of spores in wet mount preparations or routine histological sections using either hematoxylin and eosin or special stains such as Fungi-Fluor (Kent & Bishop-Stewart 2003), acid fast (Ramsay et al. 2009), or Luna (Peterson et al. 2011). The development of a conventional PCR based test (Whipps & Kent 2006) has allowed researchers to detect lower levels of the parasite in fish tissues than possible by traditional histological methods and was used effectively to screen brood fish and progeny to establish a specific pathogen free (SPF) zebrafish colony (Kent et al. 2009, Kent et al. in press). These testing methods are relatively sensitive and specific, but were not evaluated in non-lethal formats. Many populations of zebrafish used in research consist of small numbers of difficult to breed mutant lines of fish and thus it is often impractical to lethally sample statistically significant numbers for diagnostic testing. For example, to screen a population of 100 fish, assuming a one or greater percent prevalence of infection in the population, 96 fish would need to be examined in order to obtain 95% confidence in detecting at the infection at a prevalence is 1% or greater (Kent et al. 2009, Simon & Schill 1984). Therefore, detection of P. neurophilia in the fish's environment or in spawning products offers a practical and desirable alternative.
Several studies have been undertaken that incorporated various techniques for concentration and detection of microparasites in water. These include membrane filtration, continuous flow centrifugation and some in combination with immunomagnetic separation or PCR to detect microsporidia and other small parasites in large volumes of drinking or surface water (Bukhari et al. 1998, Fournier et al. 2000, Swales & Wright 2000, Hallett & Bartholomew 2006, Graczyk et al. 2007, Hoffman et al. 2007). Graczyk et al (1997) described the use of cellulose-acetate membrane filtration of water followed by dissolution of the filter using acetone to concentrate and visualize Cryptosporidium oocysts. This approach for spore concentration provided a sensitive method for detecting P. neurophilia in water.
Successful detection of microsporidia in environmental samples using PCR also requires efficient extraction of DNA from spores. A large portion of our study was, therefore, devoted to determination of the method that yields the highest amount of PCR product from spores. Both extraction comparison trials, each utilizing different sample handling, showed that sonication increased the overall sensitivity of the test. In the initial DNA extraction comparison, samples were pretreated in pools and aliquots were taken from these pools for purification in order to minimize the effects of sampling error inherent in dealing with such a small number of spores. We have found it occasionally difficult to obtain homogenous suspensions of P. neurophilia spores and this is likely attributable to some non-polar factors on the exterior of the spore wall, as we often find the spores clustered in wet mount preparations, particularly around the meniscus of air bubbles. The second DNA extraction comparison was based upon the results of the first trial and further confirmed that sonication resulted in much greater DNA extracted even taking into account potential sampling errors. Our results were consistent with those of Phelps and Goodwin (2007), who showed that the amount of DNA obtained from another egg-associated fish microsporidium, Ovipleistophora ovariae, was increased over 500 times by sonicatingspores compared to proteinase K digestion alone. As with their study, we observed numerous intact spores in preparations following the QIAgen extraction, indicating that this method and others which depend solely on proteinase K and detergent digestion is not effective for disrupting some microsporidian spores. This is further supported by the higher overall sensitivity obtained from tissue samples which had been sonicated prior to DNA extraction.
Although the extraction comparison showed that sonication without the additional step of DNA extraction using the QIAgen kit had the highest sensitivity with purified spores, this step was necessary for extraction from tank water and eggs due to inhibitory elements found in these sample matrices as determined by spiking sonicated, unpurified samples (data not shown). Whereas this inhibition could potentially be eliminated by diluting the samples or adding adjuncts such as bovine serum albumin, the DNA extraction step was added in order to maintain a consistent extraction with samples potentially containing variable amounts of inhibitory substances.
Our test was also effective for spawning water, eggs and sperm. We were able to detect very low levels of the parasite in replicates within trials and obtained similar results with independent trials. Zebrafish are spawned by placing pairs or groups into small tanks with screen bottoms overnight. Using the F1 progeny of surface-disinfected eggs is the main source for establishing new populations of zebrafish and introducing new lines into existing zebrafish colonies (Lawrence 2007), an approach that has been used for decades to avoid movement of salmonid pathogens (Stead & Laird 2002, Kent & Kieser 2003). For the latter, sex products are also screened for specific pathogens that are maternally transmitted (Miriam et al. 1997). The use of surface disinfected eggs is likely to be effective for avoiding certain bacterial pathogens but this practice has not stopped the spread of Pseudoloma neurophilia within zebrafish colonies. The reason for this is likely twofold; the ineffectiveness of levels of chlorine used for disinfection of eggs (Ferguson et al 2007) and the maternal transmission route of the parasite. Other fish microsporidia are vertically transmitted (Phelps & Goodwin 2008), and several lines of indirect and observational data indicate that there is a risk of maternal transmission with P. neurophilia, either transovum (within eggs) or transovarial (outside egg at spawning): 1) the parasite has been observed in several facilities that have been established and only use F1 progeny from disinfected eggs, 2) the parasite is common in the ovaries and occasionally within eggs, 3) larval zebrafish are highly susceptible to the infections. We detected the parasite in water and eggs from a spawn tank of a population of brood fish with an infection prevalence of 60%, providing further evidence of maternal transmission.
Given that resistant spores are abundant at spawning, the only way to reliably avoid maternal transmission is to identify infected broodfish or progeny with a highly sensitive diagnostic test. Screening of 10 day old fry and broodstock from several zebrafish lines were used to establish a P. neurophilia SPF facility at Oregon State University (Kent et al. 2009, Kent et al In Press). Our test also has potential to be used for nonlethal screening of adult stocks. This non-lethal format is feasible as adult zebrafish spawn prodigiously and paired spawning is often performed in 1 l or less of water (Lawrence 2007, Harper & Lawrence 2011). The results of the group spawn experiment illustrate the potential application of this assay in this format. However, it is important to note that testing sex products and water may not be reliable for detecting the parasite in all infected fish. The parasite does not occur in the ovaries of all infected females zebrafish (Kent & Bishop-Stewart 2003) and thus these fish may be less likely to shed spores at spawning. Also, we have not seen the parasite in the testis by histology, and thus this approach for non-lethal testing may not be useful for identifying infected males.
With these limitations, the test would still be of value in that a positive result from pooled samples from a group spawn would clearly demonstrate that at least one fish in the spawning group was positive. Moreover, our test is extremely sensitive, and thus a negative result with spawning water or eggs would be highly suggestive that the progeny from this particular spawn were not infected. Other experiments performed in our laboratory have produced similar results and further studies are currently underway to elucidate the progression and transmission of the parasite within populations of fish in order to determine the predictive value of this type of test and its reliability in detecting positive fish in lightly infected populations. We are also further investigating the role of males in maternal transmission of the parasite. Cryopreserved sperm is used to for long term storage of zebrafish lines and is frequently used to establish new populations (Westerfield 2007). Whereas we have not detected the parasite in testis by histology, sperm could be contaminated during the squeezing process to obtain sperm. Hence, our new test provides a method for screening sperm for the parasite.
In conclusion, we provide here a new real-time PCR based assay for P. neurophilia that is more sensitive that our previous test. Additionally, we have developed a method by which to sample and detect the parasite in water, eggs, and sperm, thus providing the foundation for a non-lethal test. It is intended that personnel involved in the maintenance of laboratory Danio rerio populations use this protocol as a basis for their own testing protocols and modify it to suit the needs of individual facilities for monitoring and screening.
Acknowledgements
This study was supported by grants from the National Institutes of Health (NIH NCRR 5R24RR017386-02 and NIH NCRR P40 RR12546-03S1). We thank Dr. C. Whipps, State University of New York, and Dr. T. Peterson, Oregon State University, for manuscript review, C. Buchner of the Sinnhuber Aquatic Research Laboratory for providing negative control specimens, and G. Weaver, Oregon State University Department of Statistics, for statistical support.
LITERATURE CITED
- Amatruda JF, Shepard JL, Stern HM, Zon LI. Zebrafish as a cancer model system. Cancer Cell. 2002;1:229–231. doi: 10.1016/s1535-6108(02)00052-1. [DOI] [PubMed] [Google Scholar]
- Baker DG. Natural pathogens of laboratory mice, rats, and rabbits and their effects on research. Clin Microbiol Rev. 1998;11(2):231–266. doi: 10.1128/cmr.11.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AMV, Kent ML. Molecular diagnostics for Loma salmonae and Nucleospora salmonis (Microsporidia). In: Cunningham C, editor. Molecular diagnosis of salmonid diseases. Kluwer Acad. Publ.; London, England: 2002. pp. 267–283. [Google Scholar]
- Bukhari Z, McCuin RM, Fricker CR, Clancy JL. Immunomagnetic separation of Cryptosporidium parvum from source water samples of various turbidities. Appl Environ Microbiol. 1998;64:4495–4499. doi: 10.1128/aem.64.11.4495-4499.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornman RS, Chen YP, Schatz MC, Street C, Zhao Y, Desany B, Egholm M, Hutchison S, Pettis JS, Lipkin WI, Evans JD. Genomic analyses of the microsporidian Nosema ceranae, an emergent pathogen of honey bees. PLoS Pathog. 2009;5:e1000466. doi: 10.1371/journal.ppat.1000466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delarbre S, Gatti S, Scaglia M, Dancourt M. Genetic diversity in the microsporidian Encephalitozoon hellem demonstrated by pulsed-field gel electrophoresis. J Eukaryot Microbiol. 2001;48:471–474. doi: 10.1111/j.1550-7408.2001.tb00181.x. [DOI] [PubMed] [Google Scholar]
- Docker MF, Devlin RH, Richard J, Khattra J, Kent ML. Sensitive and specific polymerase chain reaction assay for detection of Loma salmonae (Microsporea). Dis Aquat Org. 1997;29:41–48. [Google Scholar]
- Dooley K, Zon LI. Zebrafish: a model system for the study of human disease. Curr Opin Genet Dev. 2000;10:252–256. doi: 10.1016/s0959-437x(00)00074-5. [DOI] [PubMed] [Google Scholar]
- Dowd SE, Gerba CP, Pepper IL. Confirmation of the human-pathogenic microsporidia Enterocytozoon bieneusi, Encephalitozoon intestinalis, and Vittaforma corneae in water. Appl Environ Microbiol. 1998;64:3332–3335. doi: 10.1128/aem.64.9.3332-3335.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen JS. Zebrafish make a big splash. Cell. 1996;87:969–977. doi: 10.1016/s0092-8674(00)81792-4. [DOI] [PubMed] [Google Scholar]
- Ferguson JA, Watral V, Schwindt AR, Kent ML. Spores of two fish microsporidia (Pseudoloma neurophilia and Glugea anomala) are highly resistant to chlorine. Dis Aquat Org. 2007;76:205. doi: 10.3354/dao076205. [DOI] [PubMed] [Google Scholar]
- Fournier S, Liguory O, Santillana-Hayat M, Guillot E, Sarfati C, Dumoutier N, Molina J, Derouin F. Detection of microsporidia in surface water: a one-year follow-up study. FEMS Immunol Med Mic. 2000;29:95–100. doi: 10.1111/j.1574-695X.2000.tb01510.x. [DOI] [PubMed] [Google Scholar]
- Graczyk TK, Cranfield MR, Fayer R. Recovery of waterborne oocysts of Cryptosporidium from water samples by the membrane-filter dissolution method. Parasitol Res. 1997;83:121–125. doi: 10.1007/s004360050221. [DOI] [PubMed] [Google Scholar]
- Graczyk TK, Sunderland D, Tamang L, Shields TM, Lucy FE, Breysse PN. Quantitative evaluation of the impact of bather density on levels of human-virulent microsporidian spores in recreational water. Appl Environ Microbiol. 2007;73:4095–4099. doi: 10.1128/AEM.00365-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallett SL, Bartholomew JL. Application of a real-time PCR assay to detect and quantify the myxozoan parasite Ceratomyxa shasta in river water samples. Dis Aquat Org. 2006;71:109. doi: 10.3354/dao071109. [DOI] [PubMed] [Google Scholar]
- Harper C, Lawrence M. The laboratory zebrafish. CRC Press; Boca Raton, FL: 2011. Breeding. pp. 47–52. [Google Scholar]
- Higes M, Martin R, Meana A. Nosema ceranae, a new microsporidian parasite in honeybees in Europe. J Invertebr Pathol. 2006;92:93–95. doi: 10.1016/j.jip.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Hill AJ, Teraoka H, Heideman W, Peterson RE. Zebrafish as a model vertebrate for investigating chemical toxicity. Toxicol Sci. 2005;86:6–19. doi: 10.1093/toxsci/kfi110. [DOI] [PubMed] [Google Scholar]
- Hoffman RM, Wolk DM, Spencer SK, Borchardt MA. Development of a method for the detection of waterborne microsporidia. J Microbiol Methods. 2007;70:312–318. doi: 10.1016/j.mimet.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Joseph J, Sharma S, Murthy SI, Krishna PV, Garg P, Nutheti R, Kenneth J, Balasubramanian D. Microsporidial Keratitis in India: 16S rRNA Gene-Based PCR Assay for Diagnosis and Species Identification of Microsporidia in Clinical Samples. Invest Ophthalmol Vis Sci. 2006;47:4468–4473. doi: 10.1167/iovs.06-0376. [DOI] [PubMed] [Google Scholar]
- Kent ML, Bishop-Stewart JK. Transmission and tissue distribution of Pseudoloma neurophilia (Microsporidia) of zebrafish, Danio rerio (Hamilton). J Fish Dis. 2003;26:423–426. doi: 10.1046/j.1365-2761.2003.00467.x. [DOI] [PubMed] [Google Scholar]
- Kent ML, Buchner C, Watral V, Sanders JL, LaDu J, Peterson TS, Tanguay R. Development and maintenance of a specific pathogen free (SPF) zebrafish research facility for Pseudoloma neurophilia. Dis Aquat Org. doi: 10.3354/dao02333. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent ML, Feist SW, Harper C, Hoogstraten-Miller S, Law JM, Sánchez-Morgado JM, Tanguay RL, Sanders GE, Spitsbergen JM, Whipps CM. Recommendations for control of pathogens and infectious diseases in fish research facilities. Comp Biochem Physiol C. 2009;149:240–248. doi: 10.1016/j.cbpc.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent ML, Kieser D. Biosecurity in Aquaculture Production Systems: Exclusion of Pathogens and Other Undesirables. The World Aquaculture Society; Baton Rouge, Louisiana: 2003. Avoiding the introduction of exotic pathogens with atlantic salmon, Salmo salar, reared in British Columbia. pp. 43–50. 70803. [Google Scholar]
- Kent ML, Spitsbergen JM, Matthews JM, Fournie JW, Westerfield M. ZIRC Health Services Zebrafish Disease Manual. [January 10, 2010];Diseases of Zebrafish in Research Facilities. 2007 Available at: http://zebrafish.org/zirc/health/diseaseManual.php.
- Kent ML, Whipps CM, Matthews JL, Florio D, Watral V, Bishop-Stewart JK, Poort M, Bermudez L. Mycobacteriosis in zebrafish (Danio rerio) research facilities. Comp Biochem Physiol C Toxicol Pharmacol. 2004;138:383–390. doi: 10.1016/j.cca.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Keohane EM, Weiss LM. The structure, function, and composition of the microsporidian polar tube. In: Wittner M, Weiss LM, editors. The microsporidia and microsporidiosis. American Society for Microbiology; Washington, DC: 1999. pp. 196–224. [Google Scholar]
- Kuehl RO. Design of experiments: Statistical principles of research design and analysis. 2nd edition Duxbury/Thomson Learning; Pacific Grove, CA: 2000. pp. 98–103. [Google Scholar]
- Lawrence C. The husbandry of zebrafish (Danio rerio): a review. Aquaculture. 2007;269:1–20. [Google Scholar]
- Lindstrom NM, Call DR, House ML, Moffitt CM, Cain KD. A quantitative enzyme-linked immunosorbent assay and filtration-based fluorescent antibody test as potential tools to screen broodstock for infection with Flavobacterium psychrophilum. J Aquat Anim Health. 2009;21:43–56. doi: 10.1577/H08-031.1. [DOI] [PubMed] [Google Scholar]
- Lom J, Dykova I. Protozoan parasites of fishes. Elsevier; Amsterdam, The Netherlands: 1992. p. 127. [Google Scholar]
- López-Vázquez C, Dopazo CP, Olveira JG, Barja JL, Bandín I. Development of a rapid, sensitive and non-lethal diagnostic assay for the detection of viral haemorrhagic septicaemia virus. J Virol Methods. 2006;133:167–174. doi: 10.1016/j.jviromet.2005.10.033. [DOI] [PubMed] [Google Scholar]
- Matthews JL. The Zebrafish: Genetics, Genomics, and Informatics. Vol. 77. Academic Press; 2004. Common Diseases of Laboratory Zebrafish. pp. 617–643. [DOI] [PubMed] [Google Scholar]
- Matthews JL, Brown AMV, Larison K, Bishop-Stewart JK, Rogers P, Kent ML. Pseudoloma neurophilia ng, n. sp., a new microsporidium from the central nervous system of the zebrafish (Danio rerio). J Eukaryot Microbiol. 2001;48:227–233. doi: 10.1111/j.1550-7408.2001.tb00307.x. [DOI] [PubMed] [Google Scholar]
- Miriam A, Griffiths SG, Lovely JE, Lynch WH. PCR and probe-PCR assays to monitor broodstock Atlantic salmon (Salmo salar L.) ovarian fluid and kidney tissue for presence of DNA of the fish pathogen Renibacterium salmoninarum. J Clin Microbiol. 1997;35:1322. doi: 10.1128/jcm.35.6.1322-1326.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller A, Stellermann K, Hartmann P, Schrappe M, Fätkenheuer G, Salzberger B, Diehl V, Franzen C. A powerful DNA extraction method and PCR for detection of microsporidia in clinical stool specimens. Clin Diagn Lab Immun. 1999;6:243–246. doi: 10.1128/cdli.6.2.243-246.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson TS, Spitsbergen JM, Feist SW, Kent ML. Luna stain, an improved selective stain for detection of microsporidian spores in histologic sections. Dis Aquat Org. 2011 doi: 10.3354/dao02346. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps NBD, Goodwin AE. Validation of a quantitative PCR diagnostic method for detection of the microsporidian Ovipleistophora ovariae in the cyprinid fish Notemigonus crysoleucas. Dis Aquat Org. 2007;76:215–21. doi: 10.3354/dao076215. [DOI] [PubMed] [Google Scholar]
- Phelps NBD, Goodwin AE. Vertical transmission of Ovipleistophora ovariae (microspora) within the eggs of the golden shiner. J Aquat Anim Health. 2008;20:45–53. doi: 10.1577/H07-029.1. [DOI] [PubMed] [Google Scholar]
- Ramsay JM, Watral V, Schreck CB, Kent ML. Pseudoloma neurophilia infections in zebrafish Danio rerio: effects of stress on survival, growth, and reproduction. Dis Aquat Org. 2009;88:69–84. doi: 10.3354/dao02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders JL, Lawrence C, Nichols DK, Brubaker JF, Peterson TS, Murray KN, Kent ML. Pleistophora hyphessobryconis (Microsporidia) infecting zebrafish Danio rerio in research facilities. Dis Aquat Org. 2010;91:47–56. doi: 10.3354/dao02245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RW, Kent ML, Adamson ML. Viability of Loma salmonae (Microsporidia) under laboratory conditions. Parasitol Res. 2000;86:978–981. doi: 10.1007/pl00008529. [DOI] [PubMed] [Google Scholar]
- Simon RC, Schill WB. Tables of sample size requirements for detection of fish infected by pathogens: three confidence levels for different infection prevalence and various population sizes. J Fish Dis. 1984;7:515–520. [Google Scholar]
- Stead SM, Laird LM. Handbook of salmon farming. Springer-Praxis; Aberdeen, UK: 2002. [Google Scholar]
- Swales C, Wright S. Evaluation of a continuous flow centrifuge for recovery of Cryptosporidium oocysts from large volume water samples. Water Res. 2000;34:1962–1966. [Google Scholar]
- Westerfield M. The zebrafish book, 5th Edition; A guide for the laboratory use of zebrafish (Danio rerio) Eugene, University of Oregon Press; Eugene, OR: 2007. [Google Scholar]
- Whipps CM, Kent ML. Polymerase chain reaction detection of Pseudoloma neurophilia, a common microsporidian of zebrafish (Danio rerio) Reared in Research Laboratories. J Am Assoc Lab Anim. 2006;45:36–39. [PMC free article] [PubMed] [Google Scholar]
- Williams BA, Lee RCH, Becnel JJ, Weiss LM, Fast NM, Keeling PJ. Genome sequence surveys of Brachiola algerae and Edhazardia aedis reveal microsporidia with low gene densities. BMC Genomics. 2008;9:200. doi: 10.1186/1471-2164-9-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Wittner W, Tanowitz HB, Kotler D, Cali A, Weiss LM. Small subunit rRNA sequence of Enterocytozoon bieneusi and its potential diagnostic role with use of the polymerase chain reaction. J Infect Dis. 1993;168:1570–1575. doi: 10.1093/infdis/168.6.1570. [DOI] [PubMed] [Google Scholar]