The biochemical basis for most of the morphological changes associated with apoptosis can be traced directly or indirectly to the actions of caspases, a family of intracellular cysteine proteases that function as effectors of programmed cell death (1, 2). Much of the recent progress toward mapping pathways for caspase activation has come from evaluations of normal dividing cells or established tumor lines, where obtaining large numbers of cells for biochemical analysis or transferring genes for functional analysis is readily possible. But, do all types of animal cells contain the same wiring instructions when it comes to connecting steps in cell suicide pathways? Researchers studying cell death in the heart are beginning to probe this question, and they are finding some surprises.
Adult cardiac myocytes are terminally differentiated, postreplicative cells whose intimate connections to each other are needed for establishing harmony of electrical conduction and contractile function. The cytosol of these highly specialized cells and their close counterparts within skeletal muscle contains probably the highest density of mitochondria of any tissues, making them unique. Hints that programmed cell death involving muscle cells may occur by routes that differ at least morphologically from other cells have come from studies of intersegmental muscle cell deaths that occur during metamorphosis of the moth, Manduca sexta (3). Moreover, cardiomyocytes do not exhibit the classical nuclear morphology associated with apoptosis in other tissues, even though DNA fragmentation occurs, which often is accepted as a sign of apoptosis (4, 5). It remains unknown whether these differences in morphology reflect fundamentally unique cell suicide programs versus tissue-specific differences in the repertoire of caspases or caspase substrates expressed in muscle cells, such as DFF45 and CIDE family proteins that release associated endonucleases on caspase cleavage (6, 7).
One of the events commonly associated with apoptosis involves the participation of mitochondria as caspase activators (8). Mitochondria release cytochrome c (Cyt c) into the cytosol, where it binds to and induces oligomerization of Apaf-1 (9–11), a mammalian homologue of the Caenorhabditis elegans cell death protein CED-4 (12). Once activated by Cyt c, oligomerized Apaf-1 then binds pro-caspase-9, resulting in pro-caspase-9 proteolytic self-processing and activation, followed by caspase-9-mediated cleavage and activation of pro-caspase-3 (13). Active caspase-3 then both directly participates as a terminal effector of apoptosis by cleaving various substrate proteins and also activates additional pro-caspases.
Evidence suggesting a potential difference in the way apoptosis pathways are regulated in adult cardiomyocytes is presented in this issue of the Proceedings by Nurula et al. (14), who studied human heart specimens derived from patients undergoing heart transplantation for ischemic or idiopathic-dilated cardiomyopathy. Similar to previous studies (15, 16), these investigators observed occasional cardiomyocytes within diseased hearts that displayed evidence of DNA fragmentation by in situ end-labeling (i.e., terminal deoxynucleotidyltransferase-mediated UTP nick end labeling) and gross nuclear morphological changes consistent with an apoptosis-like process. However, although only occasional cells exhibited obvious signs of deterioration, Cyt c release from mitochondria and proteolytic processing of pro-caspase-3 were readily detectable by immunoblot analysis of tissue extracts, implying that mitochondrial permeability barrier function and caspase activation had occurred in a substantial proportion of the cells within these diseased hearts. Moreover, most of the PKCδ, a known substrate of caspase-3 (17), was proteolytically processed, further suggesting a wide-scale activation of caspases that could not be limited to a few cells. Furthermore, immunoelectron microscopy revealed redistribution of Cyt c from mitochondria to cytosol in cardiomyopathy specimens but not healthy hearts derived from accident victims. The majority of these same cells with cytosolic Cyt c, however, had no evidence of the ultrastructural changes typically associated with caspase activation and apoptosis.
Interrogating the pathobiochemical events associated with cardiomyopathy in humans is difficult, given the delays typically involved during the postharvesting period. Thus, interpretation of any results derived from human heart specimens comes with caveats. Assuming, however, that Cyt c release and caspase-3 processing did occur in vivo, the question is why do the vast majority of these cardiomyocytes appear morphologically normal without evidence of apoptosis?
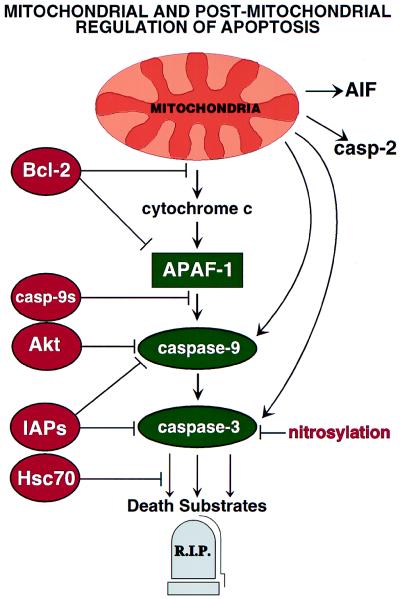
Several examples of postmitochondrial blocks to apoptosis have been delineated recently that might account for the observations of Narula et al. (14) (Fig. 1). For example, some IAP (inhibitor of apoptosis protein) family proteins have been reported to directly inhibit caspase-3 and certain caspases downstream of Cyt c (18–20). Though Narula et al. found evidence of processed caspase-3 in heart extracts, protease activity was not measured, thus it is possible that active caspase-3 was sequestered in a complex with IAP-family proteins. In addition, caspases may be inactivated by S-nitrosylation of their active site cysteine residues (21, 22), a reasonable possibility given that patients with ischemic cardiomyopathy often are treated with nitroglycerin or related nitrates. However, the concomitant presence in heart extracts of a processed caspase-3 substrate, PKCδ, argues otherwise.
Figure 1.
Postmitochondrial regulation of apoptosis. Mitochondria have been reported to sequester and release several proteins implicated in apoptosis, including Cyt c, pro-caspases-2, -3, and -9, and AIF, a flavoprotein that induces DNA degradation and other manifestations of nuclear apoptosis (60). Several steps within the Cyt c pathway may be subject to suppression by anti-apoptotic proteins (red).
Proteolytic processing of pro-caspase-3 can occur as a postlysis artifact in some tissues (23), which raises issues about the in vivo status of caspase-3 in failing hearts, but does not explain why cardiomyocytes seemingly tolerate cytosolic Cyt c (14). Several ways of preventing Cyt-c-induced caspase activation have been demonstrated (Fig. 1). For example, phosphorylation of human pro-caspase-9 can suppress Cyt-c-induced processing of pro-caspase-9 and inhibit active caspase-9 (24). Among the kinases involved in this phosphorylation is Akt (PKB), which is implicated in cell survival signaling by various growth factor receptors via the phosphatidylinositol 3′ kinase pathway (25). In addition, an isoform of pro-caspase-9 produced through alternative mRNA splicing is found in some tissues. The shorter pro-caspase-9S protein retains the N-terminal CARD domain required for interacting with Apaf-1, but lacks a region required for catalytic activity, functioning as a transdominant competitor of the full-length pro-caspase-9 protein (26, 27).
Another way to create resistance to Cyt c may be to sequester pro-caspase-9 in noncytosolic compartments. Pro-caspase-9 resides predominantly inside mitochondria in some tissues in vivo, including cardiomyocytes, skeletal muscle, and neurons, becoming released during apoptosis (28, 29). Thus, to the extent that pro-caspase-9 is unavailable in the cytosol because of mitochondrial sequestration, cells could be rendered insensitive to the presence of cytosolic Cyt c, given that pro-caspase-9 is the only caspase presently known to be a target of Apaf-1. Conceivably, postreplicative cells such as adult cardiomyocytes and differentiated neurons might have developed mechanisms for intramitochondrial sequestration of pro-caspase-9 as a way of reducing chances of apoptosis caused by inappropriate activation of Apaf-1 through non-Cyt c-dependent mechanisms or through Apaf-1-independent activation of pro-caspase-9 by cytosolic proteins such as Bcl-10 (mammalian E10) (30), a CARD-domain protein that binds and triggers activation of pro-caspase-9 when overexpressed in cell (31). Though uncertain as to its relevance, small proportions of total cellular pro-caspase-2 and pro-caspase-3 also reportedly reside in the intermembrane space of mitochondria, undergoing release into the cytosol during mitochondrial stress (29, 32).
A simple method of creating postmitochondrial resistance to Cyt c-mediated caspase activation is simply to not express Apaf-1, as has been suggested recently for skeletal muscle (33). But, what if Apaf-1 is present? Some Bcl-2 family antiapoptotic proteins may bind Apaf-1 and inhibit its function. For example, Bcl-XL reportedly forms complexes with Apaf-1 and may inhibit Apaf-1-mediated processing of pro-caspase-9, analogously to the C. elegans proteins CED-9 (Bcl-2 homolog) and CED-4 (Apaf-1 homolog) (34–37). Moreover, the Bcl-2 family member Boo/Diva also binds Apaf-1, and in some contexts may suppress apoptosis while in others promoting cell death (38, 39). Although it has been difficult to show evidence that Bcl-2 interacts with Apaf-1, in some circumstances overexpression of Bcl-2 nevertheless prevents caspase activation induced by microinjection of Cyt c (40, 41). In a cell-free system lacking mitochondria, addition of Bcl-2 protein also reportedly increased the threshold amount of Cyt c required to trigger caspase activation (42). Thus, suppression of Apaf-1 by antiapoptotic Bcl-2 family proteins could conceivably explain why cardiomyocytes of patients with end-stage heart failure seemingly tolerate Cyt c in their cytosol.
Bcl-2 and Bcl-XL also inhibit Cyt c release from mitochondria, thus accounting for much of their antiapoptotic effects in cells (8, 43, 44) (Fig. 1). This function of Bcl-2 and Bcl-XL is caspase independent and may be related to their structural similarity to certain pore-forming proteins that are known to exist in both soluble and membrane-inserted conformations (8, 45, 46). Interestingly, though Cyt c was reportedly found largely in the cytosol of cardiomyocytes of heart failure patients by Narula et al. (14), transmission electron microscopy analysis suggested that the mitochondria were normal from a ultrastructural standpoint. One must presume, therefore, that Cyt c release occurred through a change in outer membrane permeability of these organelles, and not as a result of the phenomenon of permeability transition (PT) where loss of inner membrane integrity results in mitochondrial swelling and secondary release of Cyt c and other proteins located in the intermembrane space caused by outer membrane rupture (8, 47). Elevations in cytosolic Ca2+ often associated with ischemia-impaired myocardium therefore are unlikely to explain the release of Cyt c, because Ca2+-induced release of Cyt c from isolated heart mitochondria involves PT induction (48). However, Ca2+-induced release of Cyt c from isolated brain mitochondria can occur without PT induction or swelling (49), making it difficult to exclude similar mechanisms in heart failure. Bax, a proapoptotic Bcl-2 family member, also can induce Cyt c release from isolated mitochondria through a swelling-independent mechanism (50). This observation may be relevant because Bax elevations in cardiomyocytes have been associated with heart failure in animal models (51, 52).
If much of the Cyt c were lost in vivo from the mitochondria of cardiomyocytes of patients with heart failure (14), thus causing a cessation in electron chain transport and oxidative phosphorylation, one must ask how do the cells continue to produce sufficient ATP for meeting their obliged contractile function and avoiding necrosis? Examples of Cyt c release from mitochondria in the absence of cell death have been achieved experimentally by using caspase-inhibitory compounds (53, 54). The implication therefore is that if the downstream consequences of Cyt c can be suppressed, then cell types that produce sufficient ATP through glycolysis can survive for several days, despite dumping most of their Cyt c into the cytosol.
What other factors might explain the presence of processed caspase-3 in cells that are not undergoing apoptosis? Overexpression of heat shock protein-70 kDa (Hsp70) has been reported to suppress apoptosis at a point downstream of caspase-3 activation (55). Interestingly, up-regulation of Hsp70 levels is associated with ischemic preconditioning in the heart and forced overexpression of Hsp70 is cytoprotective in cardiomyocytes (56). Moreover, overexpression of certain cochaperones such as BAG-1, a Hsp70 binding protein, prevents apoptosis in some circumstances (57, 58).
Finally, ATP depletion could explain the absence of nuclear manifestations of apoptosis despite Cyt c release in cardiomyopathy specimens. The activation of pro-caspase-9 by Cyt c and Apaf-1 depends on ATP or dATP, which binds Apaf-1 and plays an essential cofactor role (11). ATP also may be required for other steps of apoptosis downstream of caspase activation, because apoptotic nuclear changes can be inhibited by ATP depletion (59). In this regard, it is formally possible that the Cyt c release and caspase-3 processing seen by Narula et al. using resected hearts from patients with cardiomyopathy but not healthy controls reflects more rapid ex vivo deterioration in the former, with periharvest or postharvest ischemia causing ATP depletion in cardiomyocytes and thereby stymieing the occurrence of a full apoptotic program.
In summary, multiple mechanisms for forestalling apoptosis downstream of mitochondrial release of apoptogenic proteins, particularly Cyt c, have been elucidated recently. Further investigations are required for determining the relative importance of these postmitochondrial mechanisms for preventing cell death in vivo within various contexts, including heart failure.
Footnotes
A commentary on this article begins on page 8144.
References
- 1.Cryns V, Yuan Y. Genes Dev. 1998;12:1551–1570. doi: 10.1101/gad.12.11.1551. [DOI] [PubMed] [Google Scholar]
- 2.Salvesen G S, Dixit V M. Cell. 1997;91:443–446. doi: 10.1016/s0092-8674(00)80430-4. [DOI] [PubMed] [Google Scholar]
- 3.Schwartz L M, Smith S W, Jones M E E, Osbourne B A. Proc Natl Acad Sci USA. 1993;90:980–984. doi: 10.1073/pnas.90.3.980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gottlieb R A, Burleson K O, Kloner R A, Babior B M, Engler R L. J Clin Invest. 1994;94:1621–1628. doi: 10.1172/JCI117504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohno M, Takemura G, Ohno A, Misao J, Hayakawa Y, Minatoguchi S, Fujiwara T, Fujiwara H. Circulation. 1998;98:1355–1357. doi: 10.1161/01.cir.98.14.1422. [DOI] [PubMed] [Google Scholar]
- 6.Liu X, Zou H, Slaughter C, Wang X. Cell. 1997;89:175–184. doi: 10.1016/s0092-8674(00)80197-x. [DOI] [PubMed] [Google Scholar]
- 7.Inohara N, Koseki T, Chen S, Wu X, Nunez G. EMBO J. 1998;17:2526–2533. doi: 10.1093/emboj/17.9.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Green D, Reed J. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 9.Liu X, Kim C N, Yang J, Jemmerson R, Wang X. Cell. 1996;86:147–157. doi: 10.1016/s0092-8674(00)80085-9. [DOI] [PubMed] [Google Scholar]
- 10.Zou H, Henzel W J, Liu X, Lutschg A, Wang X. Cell. 1997;90:405–413. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]
- 11.Zou H, Li Y, Liu X, Wang X. J Biol Chem. 1999;274:11549–11556. doi: 10.1074/jbc.274.17.11549. [DOI] [PubMed] [Google Scholar]
- 12.Yuan J Y, Horvitz H R. Dev Biol. 1990;138:33–41. doi: 10.1016/0012-1606(90)90174-h. [DOI] [PubMed] [Google Scholar]
- 13.Li P, Nijhawan D, Budihardjo I, Srinivasula S, Ahmad M, Alnemri E, Wang X. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 14.Narula J, Pandey P, Arbustini E, Haider N, Narula N, Kolodgie F D, Bello B D, Semigran M J, Bielsa-Masdeu A, Dec G W, et al. Proc Natl Acad Sci USA. 1999;96:8144–8149. doi: 10.1073/pnas.96.14.8144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Narula J, Haider N, Virmani R, DiSalvo T, Kododgie F, Hajjar R, Schmidt U, Semigran M, Dec G, Khaw B. N Engl J Med. 1996;335:1182–1189. doi: 10.1056/NEJM199610173351603. [DOI] [PubMed] [Google Scholar]
- 16.Olivetti G, Abbi R, Quaini F, Kajstura J, Cheng W, Nitahara J A, Quaini E, Di Loreto C, Beltrami C A, Krajewski S, et al. N Eng J Med. 1997;336:1131–1141. doi: 10.1056/NEJM199704173361603. [DOI] [PubMed] [Google Scholar]
- 17.Ghayur T, Hugunin M, Talanian R V, Ratnofsky S, Quinlan C, Emoto Y, Pandey P, Datta R, Huang Y, Kharbanda S, et al. J Exp Med. 1996;184:2399–2404. doi: 10.1084/jem.184.6.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deveraux Q L, Roy N, Stennicke H R, Van Arsdale T, Zhou Q, Srinivasula M, Alnemri E S, Salvesen G S, Reed J C. EMBO J. 1998;17:2215–2223. doi: 10.1093/emboj/17.8.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deveraux Q, Takahashi R, Salvesen G S, Reed J C. Nature (London) 1997;388:300–303. doi: 10.1038/40901. [DOI] [PubMed] [Google Scholar]
- 20.Roy N, Deveraux Q L, Takashashi R, Salvesen G S, Reed J C. EMBO J. 1997;16:6914–6925. doi: 10.1093/emboj/16.23.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, Billiar T R, Talanian R V, Kim Y M. Biochem Biophys Res Commun. 1997;240:419–424. doi: 10.1006/bbrc.1997.7672. [DOI] [PubMed] [Google Scholar]
- 22.Mannick J B, Hausladen A, Liu L M, Hess D T, Zeng M, Miao Q X, Kane L S, Gow A J, Stamler J S. Science. 1999;284:651–654. doi: 10.1126/science.284.5414.651. [DOI] [PubMed] [Google Scholar]
- 23.Zapata J, Takahashi R, Salvesen G, Reed J. J Biol Chem. 1998;273:6916–6920. doi: 10.1074/jbc.273.12.6916. [DOI] [PubMed] [Google Scholar]
- 24.Cardone M, Roy N, Stennicke H, Salvesen G, Franke T, Stanbridge E, Frisch S, Reed J. Science. 1998;282:1318–1320. doi: 10.1126/science.282.5392.1318. [DOI] [PubMed] [Google Scholar]
- 25.Marte B M, Downward J. Trends Biochem Sci. 1997;22:355–358. doi: 10.1016/s0968-0004(97)01097-9. [DOI] [PubMed] [Google Scholar]
- 26.Seol D W, Billiar T R. J Biol Chem. 1999;274:2072–2076. doi: 10.1074/jbc.274.4.2072. [DOI] [PubMed] [Google Scholar]
- 27.Srinivasula S M, Ahmad M, Guo Y, Zhan Y, Lazebnik Y, Fernandes-Alnemri T, Alnemri E S. Cancer Res. 1999;59:999–1002. [PubMed] [Google Scholar]
- 28.Krajewski S, Krajewska M, Ellerby L M, Welsch K, Xie Z, Deveraux Q L, Salvesen G S, Bredesen D E, Rosenthal R E, Fiskum G, Reed J C. Proc Natl Acad Sci USA. 1999;96:5752–5757. doi: 10.1073/pnas.96.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Susin S A, Lornzo H K, Zamzami N, Marzo I, Brenner C, Larochette N, Prevost M-C, Alzari P M, Kroemer G. J Exp Med. 1999;189:382–394. doi: 10.1084/jem.189.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Willis T, Jadayel D, Peng H, Perry A, Rauf-Abdul M, Price H, Karran L, Majekodunmi O, Wlodarska I, Pan L, et al. Cell. 1999;96:35–45. doi: 10.1016/s0092-8674(00)80957-5. [DOI] [PubMed] [Google Scholar]
- 31.Yan M, Lee J, Schilbach S, Goddard A, Dixit V. J Biol Chem. 1999;275:10287–10292. doi: 10.1074/jbc.274.15.10287. [DOI] [PubMed] [Google Scholar]
- 32.Mancini M, Nicholson D, Roy S, Thornberry N, Peterson E, Casciola-Rosen L, Rosen A. J Cell Biol. 1998;140:1485–1495. doi: 10.1083/jcb.140.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burgess D, Svensson M, Dandrea T, Gröulund K, Hammarquist F, Orrenius S, Cotgreave I. Oncogene. 1999;6:256–261. doi: 10.1038/sj.cdd.4400489. [DOI] [PubMed] [Google Scholar]
- 34.Spector M S, Desnoyers S, Heoppner D J, Hengartner M O. Nature (London) 1997;385:653–656. doi: 10.1038/385653a0. [DOI] [PubMed] [Google Scholar]
- 35.Chinnaiyan A M, O’Rourke K, Lane B R, Dixit V M. Science. 1997;275:1122–1126. doi: 10.1126/science.275.5303.1122. [DOI] [PubMed] [Google Scholar]
- 36.Wu D, Wallen H D, Nunez G. Science. 1997;275:1126–1129. doi: 10.1126/science.275.5303.1126. [DOI] [PubMed] [Google Scholar]
- 37.Metzstein M, Stanfield G, Horvitz H. Trends Genet. 1998;14:410–416. doi: 10.1016/s0168-9525(98)01573-x. [DOI] [PubMed] [Google Scholar]
- 38.Song Q, Kuang Y, Dixit V M, Vincenz C. EMBO. 1999;18:167–178. doi: 10.1093/emboj/18.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Inohara N, Gourley T S, Carrio R, Muniz M, Merino J, Garcia I, Koseki T, Hu Y, Chen S, Nunez G. J Biol Chem. 1998;273:32479–32486. doi: 10.1074/jbc.273.49.32479. [DOI] [PubMed] [Google Scholar]
- 40.Rossé T, Olivier R, Monney L, Rager M, Conus S, Fellay I, Jansen B, Borner C. Nature (London) 1998;391:496–499. doi: 10.1038/35160. [DOI] [PubMed] [Google Scholar]
- 41.Zhivotovsky B, Orrenius S, Brustugun O T, Doskeland S O. Nature (London) 1998;391:441–442. doi: 10.1038/35060. [DOI] [PubMed] [Google Scholar]
- 42.Cosulich S C, Savory P J, Clarke P R. Curr Biol. 1999;9:147–150. doi: 10.1016/s0960-9822(99)80068-2. [DOI] [PubMed] [Google Scholar]
- 43.Yang J, Liu X, Bhalla K, Kim C N, Ibrado A M, Cai J, Peng I-I, Jones D P, Wang X. Science. 1997;275:1129–1132. doi: 10.1126/science.275.5303.1129. [DOI] [PubMed] [Google Scholar]
- 44.Kluck R M, Bossy-Wetzel E, Green D R, Newmeyer D D. Science. 1997;275:1132–1136. doi: 10.1126/science.275.5303.1132. [DOI] [PubMed] [Google Scholar]
- 45.Schendel S, Montal M, Reed J C. Cell Death Differ. 1998;5:372–380. doi: 10.1038/sj.cdd.4400365. [DOI] [PubMed] [Google Scholar]
- 46.Muchmore S W, Sattler M, Liang H, Meadows R P, Harlan J E, Yoon H S, Nettesheim D, Changs B S, Thompson C B, Wong S, et al. Nature (London) 1996;381:335–341. doi: 10.1038/381335a0. [DOI] [PubMed] [Google Scholar]
- 47.Susin S, Zamzami N, Kroemer G. Biochim Biophys Acta. 1998;1366:151–165. doi: 10.1016/s0005-2728(98)00110-8. [DOI] [PubMed] [Google Scholar]
- 48.Borutaite V, Morkuniene R, Brown G C. Biochim Biophys Acta. 1999;1453:41–48. doi: 10.1016/s0925-4439(98)00082-9. [DOI] [PubMed] [Google Scholar]
- 49.Andreyev A, Auldrige M, Rosenthal R E, Haywood Y, Silbergleit R, Fiskum G. FEBS Lett. 1998;439:373–376. doi: 10.1016/s0014-5793(98)01394-5. [DOI] [PubMed] [Google Scholar]
- 50.Jurgensmeier J M, Xie Z, Deveraux Q, Ellerby L, Bredesen D, Reed J C. Proc Natl Acad Sci USA. 1998;5:4997–5002. doi: 10.1073/pnas.95.9.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fortuno M A, Ravassa S, Etayo J C, Diez J. Hypertension. 1998;32:280–286. doi: 10.1161/01.hyp.32.2.280. [DOI] [PubMed] [Google Scholar]
- 52.Leri A, Claudio P, Li Q, Wang X, Reiss K, Wang S, Malhotra A, Kajstura J, Anversa P. J Clin Invest. 1998;101:1326–1342. doi: 10.1172/JCI316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen Q, Takeyama N, Brady G, Watson A J M, Dive C. Blood. 1998;92:545–4553. [PubMed] [Google Scholar]
- 54.Martinou I, Desagher S, Eskes R, Antonsson B, André E, Fakan S, Martinou J-C. J Cell Biol. 1999;144:883–889. doi: 10.1083/jcb.144.5.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jäättelä M, Wissing D, Kokholm K, Kallunki T, Egeblad M. EMBO J. 1998;17:6124–6134. doi: 10.1093/emboj/17.21.6124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Benjamin I J, McMillan R D. Circ Res. 1998;83:117–132. doi: 10.1161/01.res.83.2.117. [DOI] [PubMed] [Google Scholar]
- 57.Takayama S, Sato T, Krajewski S, Kochel K, Irie S, Millan J A, Reed J C. Cell. 1995;80:279–284. doi: 10.1016/0092-8674(95)90410-7. [DOI] [PubMed] [Google Scholar]
- 58.Takayama S, Bimston D N, Matsuzawa S, Freeman B C, Aime-Sempe C, Xie Z, Morimoto R J, Reed J C. EMBO J. 1997;16:4887–4896. doi: 10.1093/emboj/16.16.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eguchi Y, Srinivasan A, Tomaselli K J, Shimizu S, Tsujimoto Y. Cancer Res. 1999;59:2174–2181. [PubMed] [Google Scholar]
- 60.Susin S, Lorenzo H, Zamzami N, Marzo I, Snow B, Brothers G, Mangion J, Jacotot E, Costantini P, Loeffler M, et al. Nature (London) 1999;397:441–446. doi: 10.1038/17135. [DOI] [PubMed] [Google Scholar]