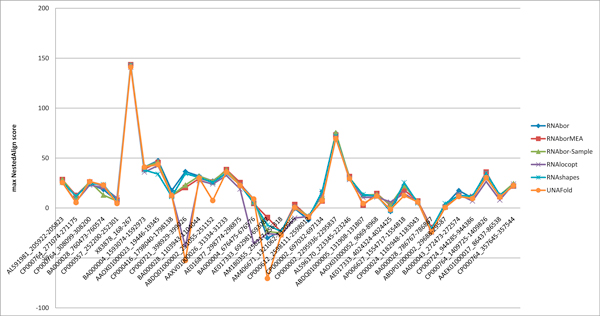
Figure 6.
For each RNA sequence in the seed alignment from Rfam family RF00167 of purine riboswitch aptamers, we retrieved downstream flanking residues from the appropriate EMBL files, in order to ensure likelihood that the expression platform was included. Then the following six programs were run: RNAbor, RNAborMEA, RNAbor-Sample, RNAlocopt, RNAshapes, UNAFold. Each program outputs a number of near-optimal secondary structures, each according to different criteria. Taking RNAbor and RNAborMEA as examples, the programs RNAbor and RNAborMEA were run, in order to compute the MFE(k) structure and the MEA(k) structure, which have minimum free energy resp. maximum expected accuracy among all k-neighbors of the intial minimum free energy structures S0. Subsequently, we applied the program NestedAlign described in [36] to compute the structural similarity between the experimentally determined GENE OFF structure for XPT guanine riboswitch of B. subtilis; i.e. the right panel of Figure 3. (Similar structures have positive scores; dissimilar structures have negative scores.) For each RNA of the seed alignment of RF00167, we determined the value k1, such the MEA(k1) structure for that RNA has the greatest structural similarity with the XPT GENE OFF structure, as determined by NestedAlign. (See the right panel of Figure 3 for the experimentally determined GENE OFF structure of XPT.) As earlier explained, we performed similar computations for the programs RNAshapes[39] and UNAFold [41], the programs RNAborMEA and RNAbor-Sample, described in this paper, and RNAbor[9] and RNAlocopt[34]. In 22 out of 34 instances, RNAborMEA produced the secondary structure most structurally similar to the experimentally determined XPT GENE OFF structure, as measured by NestedAlign.