Fig. 2.
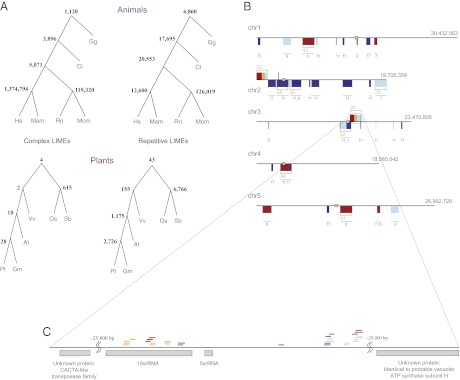
Plant LIMEs are remarkably diverse in their structure and function. (A) Phylogenetic trees of the six animal and six plant species for complex and repetitive LIMEs. Mam corresponds to Macaca mulatta, and Mum corresponds to Mus musculus. A node number (bold) is the number of elements common to each species in a subtree below. All LIMEs ≥100 bp are considered for each subtree. At, Arabidopsis thaliana; Gm, Glycine max (soybean); Hs, Homo sapiens (human); Mam, Macaca mulatta (macaque); Mum, Mus musculus (mouse); Os, Oryza sativa (rice); Pt, Populus trichocarpa (cottonwood); Rn, Rattus norvegicus (rat); Sb, Sorghum bicolor (sorghum); Vv, Vitis vinifera (grape). (B) LIMEs in the Arabidopsis (At) genome, depicted as colored ticks with complex LIMEs above and repetitive LIMEs below each chromosome (chr) sequence. Tick color corresponds to the number of genomes, including the At genome, sharing a LIME: red for three genomes, orange for four, light blue for five, and dark blue for six. When two LIMEs are 45 kbp or less apart, they are grouped in the same box. Once there are more than 20 LIMEs in such a box, the box size is unchanged but correct proportions of LIMEs shared by three, four, five, and six genomes are depicted by the relative thickness of the colored parts. Orange numbers specify the total number of LIMEs per box, and blue corresponds to the motif ID for one or multiple repetitive LIMEs. Identified centromere positions are shown as gray boxes. (C) Detailed representation of a chromosome 3 region that includes 2 LIMEs shared by all six genomes, and the nearest genes.