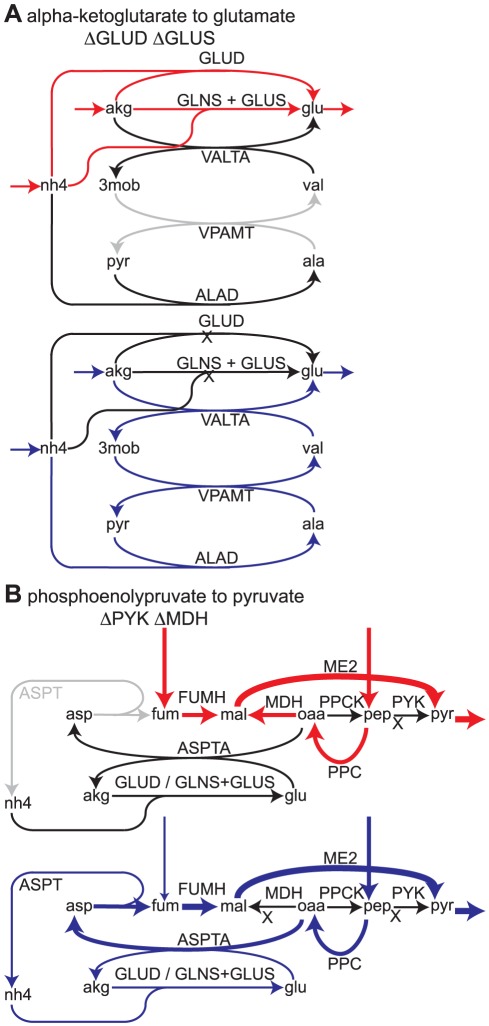
Figure 5. Identified metabolic differences in cyanobacteria.
(A) Top: Pathways for synthesis of glutamate (glu) from alpha-ketoglutarate (akg) used in iCce806. Bottom: Pathway predicted by the iSyp611 model when glutamate dehydrogenase (GLUD) and glutamate synthase (GLUS) are deleted. Valine aminotransferase (VPAMT) enables the synthesis of glutamate from pyruvate (pyr). (B) Top: Pathway for conversion of phosphoenolpyruvate (pep) to pyruvate when pyruvate kinase (PYK) is deleted from iCce806. Bottom: Pathway predicted by the iSyp611 model when malate dehydrogenase (MDH) is also deleted. Aspartase (ASPT) allows malate (mal) to be synthesized entirely from fumarate (fum), rather than from fumarate and oxaloacetate (oaa). (A and B) Red arrows indicate flux in the iCce806 model. Blue arrows represent flux in the iSyp611 model under the indicated knockout condition. Black arrows indicate inactive reactions and reaction deletions are indicated by black ‘X’s. Gray arrows (top panels) indicate reactions not present in the iCce806 model. Arrow thickness corresponds to relative flux levels. Reaction and metabolite abbreviations are identical in the iSyp611 and iCce806 models and are given in Dataset S2.