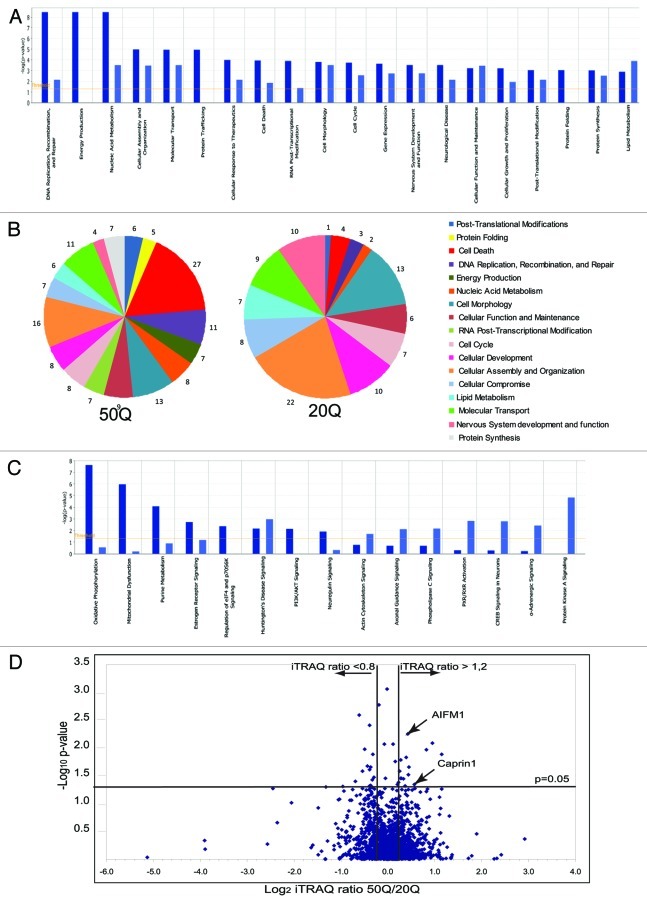
Figure 2. Quantitative analysis of normal and expanded Htt interactome. (A) Top cellular functions (as defined by IPA) most significantly enriched within Htt-50Q (dark blue) and Htt-20Q (light blue) preferential interactors, based on the protein data set shown in Table S1 (CV < 0.35 between duplicate samples). The graph shows the enrichment of particular functional categories based on the observed number of proteins for each group, relative to the number expected by chance. Log10 p-values are calculated by IPA. (B) The pie diagram, showing selected functional categories most significantly enriched within Htt-20Q and Htt-50Q preferential interactors, based on the IPA analysis of differential interactors with CV < 0.25 between duplicate samples. The numbers represent the percentage of differential interactors involved in a specific function, relative to the number of all differential interactors combined identified for all functional categories shown. (C) Top Canonical pathways (as defined by IPA) most significantly enriched within Htt-50Q (dark blue) and Htt-20Q (light blue) preferential interactors using the protein data set shown in Table S1. The graph shows the enrichment of particular canonical pathways based on the observed number of proteins for each pathway, relative to the number expected by chance. Log10 p-values are calculated by IPA. (D) Volcano plot showing the distribution of proteins differentially interacting with normal (20Q) and expanded (50Q) Htt based on iTRAQ ratios and p-values. The x-axis shows the Log2 of the median normalized iTRAQ ratios between 50Q and 20Q. The vertical lines indicate the threshold iTRAQ ratios required for a protein to be considered a differential interactor (> 1.2 or < 0.8).The x-axis shows the –Log10 of p-values, obtained using ANOVA approach, described in the Results. The horizontal line represents p-value of 0.05.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.