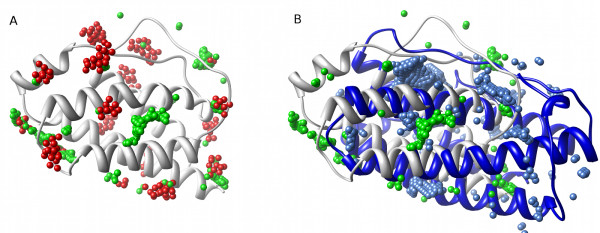
Figure 1.
The difficulties of global comparison of pockets on protein surfaces. (A) Pocket predictions from PASS (red) and LIGSITE-cs (green) for the same interleukin-2 (IL-2) structure (ribbon, PDB:1M47). The similarities and differences between these predictions are readily visualized, but how can identification of common features be automated? (B) Homologous structures: The surface pockets of IL-2 (white ribbon, green spheres) [PDB:1M47] and its distant homologue leukaemia inhibitory factor (LIF) (blue ribbon, blue spheres) [PDB:1LKI], both represented by LIGSITE-cs spheres, show little overlap. Is this because the pockets are genuinely unrelated, or because of the difficulty of making a 'correct' structural alignment?.