Abstract
Aspergillus fumigatus is a primary and opportunistic pathogen, as well as a major allergen, of mammals. The Ca+2-calcineurin pathway affects virulence, morphogenesis and antifungal drug action in A. fumigatus. Here, we investigated three components of the A. fumigatus Ca+2-calcineurin pathway, pmcA,-B, and -C, which encode calcium transporters. We demonstrated that CrzA can directly control the mRNA accumulation of the pmcA-C genes by binding to their promoter regions. CrzA-binding experiments suggested that the 5′-CACAGCCAC-3′ and 5′-CCCTGCCCC-3′ sequences upstream of pmcA and pmcC genes, respectively, are possible calcineurin-dependent response elements (CDREs)-like consensus motifs. Null mutants were constructed for pmcA and -B and a conditional mutant for pmcC demonstrating pmcC is an essential gene. The ΔpmcA and ΔpmcB mutants were more sensitive to calcium and resistant to manganese and cyclosporin was able to modulate the sensitivity or resistance of these mutants to these salts, supporting the interaction between calcineurin and the function of these transporters. The pmcA-C genes have decreased mRNA abundance into the alveoli in the ΔcalA and ΔcrzA mutant strains. However, only the A. fumigatus ΔpmcA was avirulent in the murine model of invasive pulmonary aspergillosis.
Introduction
Calcium ions are extremely important for signal transduction. Two important calcium mediators in the eukaryotic cell are calmodulin and the phosphatase calcineurin [1], [2]. Calcineurin is a heterodimeric protein composed by a catalytic subunit A and a regulatory subunit B [1]. In fungi, calcineurin plays an important role in the control of cell morphology and virulence [1], [2], [3], [4]. The main mode of action of calcineurin is through the dephosphorylation of the transcription factor Crz1p [5]. Calcineurin dephosphorylates Crz1p upon an increase in cytosolic calcium, allowing its nuclear translocation [5], [6]. CRZ1 deficient mutants display hypersensitivity to chloride and chitosan, a defective transcriptional response to alkaline stress and defects in cellular morphology and mating [5], [7], [8], [9]. Inactivated Schizosaccharomyces pombe CRZ1 mutants (Δprz1) are hypersensitive to calcium and have decreased transcription of the Pmc1 Ca+2 pump [10]. C. albicans homozygotes crz1Δ/Δ display moderately attenuated virulence and sensitive to calcium, lithium, manganese, and sodium dodecyl sulfate [6], [11], [12].
We and others have been characterizing the Ca+2-calcineurin pathway in the human pathogenic fungus A. fumigatus [3]. In this fungus calcineurin is need for hyphal extension, branching and conidial architecture. Furthermore, the A. fumigatus ΔcalA mutant strain has decreased fitness in a low dose murine infection, cannot grow in fetal bovine serum (FBS), and is deficient in inorganic phosphate transport [3], [13]. Three other elements in this pathway were also characterized: (i) the transcription factor CrzA [14], [15], (ii) the RcnA/CbpA, belonging to a class of endogenous calcineurin regulators, calcipressins [16], [17], and (iii) the Golgi apparatus Ca+2/Mn+2 P-type ATPase PmrA [16]. CrzA mediates cellular tolerance to increased concentrations of calcium and manganese [14], [15]. In addition to acute sensitivity to these ions and decreased conidiation, the crzA null mutant suffers from decreased expression of calcium transporters under high calcium concentrations and a loss of virulence. The last identified component of the pathway in A. fumigatus, PmrA, has been demonstrated to play a role in cation homeostasis and in the cell wall integrity pathway [16].
Fungal vacuolar Ca2+ ATPases are involved in removing Ca2+ ions from the cytosol and transporting them to internal stores thus avoiding calcium toxicity [18]). In fungi, the vacuole is a major calcium store and the two main pathways that facilitate the accumulation of Ca+2 into vacuoles are the Ca+2-ATPases and Ca+2/H+ exchangers [18]. In S. cerevisiae, PMC1 is responsible for this process preventing growth inhibition by the activation of calcineurin in the presence of elevated calcium concentrations [19]. Here, we report the molecular characterization of three A. fumigatus PMC1 calcium transporter-encoding genes, pmcA-C. We demonstrated that CrzA directly controls the pmcA-C mRNA accumulation via binding to their promoter regions. We constructed null mutants for pmcA-B, a conditional mutant for pmcC and investigated the phenotypes/virulence of these deletions in a murine model of invasive pulmonary aspergillosis. We show that A. fumigatus pmcC is an essential gene, while pmcA and pmcB are both involved in calcium and manganese metabolism. However, only pmcA had a dramatic impact on A. fumigatus virulence and pathogenicity, since A. fumigatus ΔpmcA was avirulent in a murine model of invasive pulmonary aspergillosis.
Results
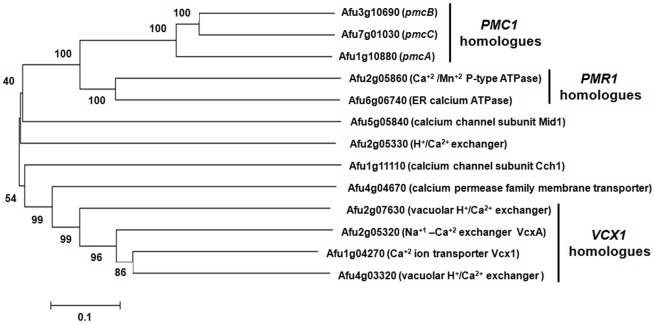
Identification of three A. fumigatus PMC1 homologues
The three main calcium transporters responsible for calcium metabolism in S. cerevisiae are PMC1, VCX1, and PMR1 [20]. A phylogenetic analysis was performed in order to learn more about homologues of these transporters and other putative A. fumigatus calcium transporters (Figure 1). Previously we observed that the mRNA abundance of two PMC1 orthologous genes, pmcA (Afu1g10880) and pmcB (Afu3g10690), which encode calcium transporters, was dependent on CalA and CrzA (Soriani et al., 2008). By using this approach, we have identified an additional PMC1 orthologue, pmcC (Afu7g01030). S. cerevisiae VCX1 encodes a vacuolar antiporter with Ca+2/H+ and K+/H+ exchange activity, which is involved in the control of cytosolic Ca+2 and K+ concentrations [21]. There are four A. fumigatus Vcx1p homologues, Afu1g04270 and Afu4g03320 (possibly paralogues), Afu2g07630 and Afu2g05320 (Figure 1). Finally, S. cerevisiae PMR1 encodes a high affinity Ca+ 2/Mn+2 P-type ATPase required for Ca+2 and Mn+2 transport into the Golgi [22]. We have identified two A. fumigatus PMR1 homologues, Afu2g05860 and Afu6g06740 (Figure 1). Recently, A. fumigatus pmrA (Afu2g05860) was characterized [16]. The ΔpmrA mutant strain has increased β-glucan and chitin content and it is hypersensitive to cell wall inhibitors, but remains virulent. In addition to these three classes of transporters, we also identified homologues for the calcium channel subunit Mid1 (Afu5g05840), an H+/Ca+2 exchanger (Afu2g05330), the calcium channel subunit Cch1 (Afu1g11110), and a calcium permease family membrane transporter (Figure 1).
Figure 1. A. fumigatus has three S. cerevisiae PMC1 homologues.
Phylogram tree and multiple sequence alignment of calcium transporter orthologues were made in CLUSTAL W2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html) using the default parameters. The followings proteins were used for the analysis: Afu3g10690 (pmcB; XP_754550); Afu7g01030 (pmcC; XP_746828); Afu1g10880 (pmcA; XP_752453); Afu2g05860 (calcium/mangenese P-type ATPase: XP_749715); Afu6g06740 (endoplasmic reticulum calcium ATPase: XP_750567); Afu5g05840 (calcium channel subunit Mid1: XP_754048); Afu2g05330 (vacuolar H+/Ca2+ exchanger: XP_749663); Afu1g11110 (calcium channel subunit Cch1: XP_752476); Afu4g04670 (calcium permease family membrane transporter: XP_746653); Afu2g07630 (vacuolar H+/Ca2+ exchanger: XP_755098); Afu2g05320 (calcium-proton exchanger: XP_749662); Afu1g04270 (calcium ion transporter Vcx1: XP_750174); and Afu4g03320 (similar to vacuolar H+/Ca2+ exchanger: XP_001481534).
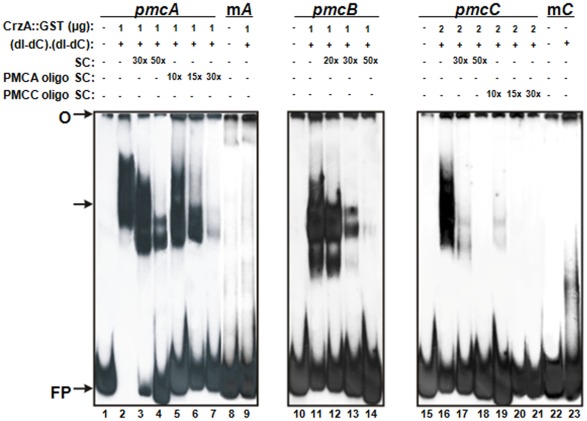
Here, we concentrate our attention on the molecular characterization of A. fumigatus PMC1 homologues. These three putative proteins showed approximately 45% identity and 67% similarity (e-value from 7.0e-160 to 1.4e-208) to the S. cerevisiae PMC1 homologue. PmcA demonstrates 51% identity and 63% similarity with PmcB (e-value 6.9e-271) and 45% identity and 58% similarity with PmcC (e-value 4.e-174) while, PmcB and PmcC showed 53% identity and 66% similarity (e-value 2.7e-276) (for the Clustal aligment of these three proteins, see Supplementary Figure S1). PmcA-C are closely related and probably paralogues (Figure 1). In addition to pmcA and pmcB, the pmcC gene also has decreased mRNA abundance in the ΔcalA and ΔcrzA mutant strains, respectively, when exposed in vitro to CaCl2 200 mM, compared to wild-type A. fumigatus [14], [17, data not shown]. To address if CrzA is directly controlling the transcription of pmcA-C, we performed Electrophoretic Mobility Shift Assays (EMSA) using purified recombinant GST::CrzA produced in E. coli. Previously, we performed an in silico analysis using MEME (Motif-based sequence analysis tools; http://meme.sdsc.edu/meme4_1_1/intro.html) to detect the possible presence of a calcineurin-dependent response elements (CDREs)-like consensus motifs in the promoter regions of 28 A. fumigatus CrzA-dependent genes [17]. By analyzing their promoter regions, 5′-GT[T/G]G[G/C][T/A]GA[G/T]-3′ was defined as the CDRE-consensus sequence for A. fumigatus AfCrzA-dependent genes. When the pmcA-C promoter regions (about 500-bp upstream ATG) were scanned for putative CDRE motifs, we were able to identify the 5′-CCCTGCCCC-3′ and 5′-CACAGCCAC-3′ sequences (at −156 and −102 bp from the ATG start codon, respectively) in the pmcA and pmcC promoter regions. However, we could not identify any conserved CDRE motif in the pmcB promoter region (Supplementary Figure S2). Three DNA fragments of about 300-bp located upstream the putative ATG initiation codon of pmcA-C genes were used as probes (Supplementary Figure S2). DNA-protein complexes with reduced mobility were observed in the three DNA fragments (Figure 2), however the complexes affinities were different among the three fragments. While 2 µg of GST::CrzA were required for the binding of the pmcC probe (Figure 2, lane 16), 1 µg of protein was enough to produce strong DNA-protein complexes for the pmcA (Figure 2, lane 2) and pmcB (Figure 2, lane 11) probes. This suggests that CrzA has low affinity to the pmcC promoter.
Figure 2. Binding of GST::CrzA recombinant protein to pmcA-C promoters.
Gel shift analysis was performed using three DNA fragments of pmcA, pmcB and pmcC promoters as probes and 1.0 µg to 2.0 µg of the CrzA::GST recombinant protein. Lanes 1 to 7, pmcA probe; lane 1, no protein added. Lanes 8 and 9, mutated pmcA probe. Lanes 10 to 14, pmcB probe; lane 10, no protein added. Lanes 11 to 14, fragments from pmcB probe. Lanes 15 to 23, pmcC probe; lane 15, no protein added. Lanes 22 and 23, mutated pmcC probe. O, gel origin; SC, specific competitor; FP, free probe. The arrow indicates the CrzA::GST-DNA complexes.
The complexes specificities were confirmed by addition of unlabelled probes as specific competitors. Addition of 50-fold molar excess of unlabelled probes completely inhibited the complexes formed with pmcB and pmcC DNA fragments. The addition of approximately a 15-fold molar excess of the DNA oligonucleotide (5′-CACAGCCAC-3′) inhibited completely the pmcC complex specificity (Figure 2, lane 20). However the complex pmcA-CrzA was only inhibited in the presence of a 30-fold molar excess of DNA oligonucleotide (5′-CCCTGCCCC-3′) containing the CDRE motif used as specific competitor (Figure 2, lane 7). This result suggests a strong CrzA affinity for this DNA fragment. The specificity of the DNA-protein complex was also confirmed by using mutated pmcA and pmcC probes, in which the core sequences were changed by site-directed mutagenesis. We have not investigated a mutated pmcB DNA fragment because we were not able to identify a conserved CDRE motif in this upstream region. We have not observed the formation of any complex by using both mutated DNA fragments as probes (Figure 2, lane 9 for mpmcA probe and lane 23 for mpmcC probe). An interesting result was the presence of two complexes exhibiting different molecular masses for pmcA and pmcB probes. We speculate that they may represent complexes with distinct conformational structures. Additional experiments will be necessary to clarify this. Taken together our results suggest that the mRNA accumulation of pmcA-C is directly regulated by CrzA.
Construction of the A. fumigatus pmcA-C mutants
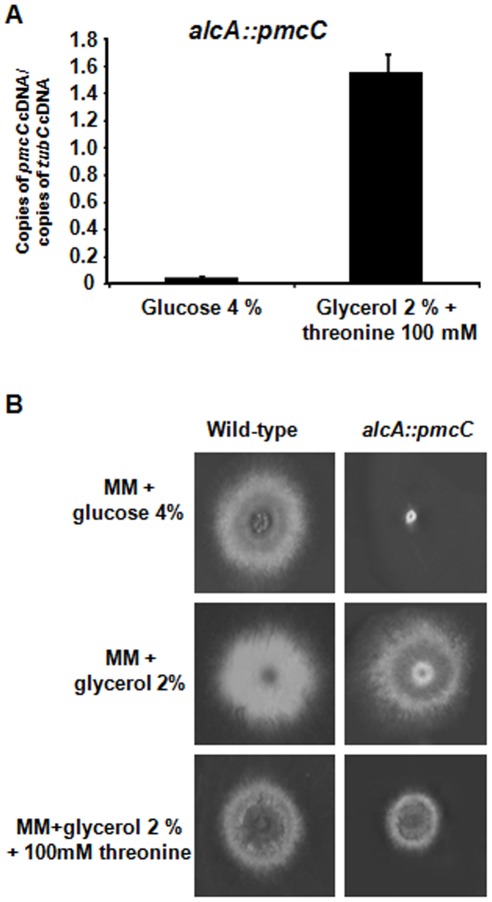
To get a greater understanding of the role of pmcA-C, we tried to inactivate all three genes (Supplementary Figure S3). However, we were unable to inactivate pmcC, suggesting that this is an essential A. fumigatus gene. Thus, we constructed an alcA::pmcC mutant by replacing the endogenous pmcC promoter with the alcA promoter and verified its growth when the alcA promoter was repressed. The alcA promoter is repressed by glucose, derepressed by glycerol and induced to high levels by ethanol or L-threonine [23]. We selected a transformant that when transferred from 16 h growth in 2% glycerol, as single carbon source, to 2% glycerol +100 mM threonine for 6 h, the mRNA accumulation of pmcC was approximately 15-fold higher than when grown in the presence of 4% glucose (Figure 3A). The repression of alcA by growing the alcA::pmcC mutant strain in the presence of 4% glucose decreased colony diameter dramatically (Figure 3B). In contrast, both wild-type and alcA::pmcC strains demonstrated similar radial diameter when grown in 2% glycerol (Figure 3B). Interestingly, pmcC overexpression also decreased the colony diameter size when compared to the wild-type strain, suggesting increased PmcC activity causes some metabolic disturbance that affects growth (Figure 3B). These results strongly indicate pmcC is an essential A. fumigatus gene.
Figure 3. The pmcC gene is an essential A. fumigatus gene.
(A) The alcA::pmcC strain was grown for 16 hours in MM+ 2% glycerol at 37°C and transferred into either MM+4% glucose or MM+2% glycerol +threonine 100 mM and grown for further 6 hours. The relative quantitation of pmcC and tubulin gene expression was determined by a standard curve (i.e., CT –values plotted against a logarithm of the DNA copy number). The results are the means (± standard deviation) of four biological replicates. (B) Growth phenotypes of the alcA::pmcC mutant strain. The A. fumigatus wild-type and alcA::pmcC mutant strains were grown for 72 hours at 37°C in MM+4% glucose, MM+2% glycerol and MM+2% glycerol +threonine 100 mM.
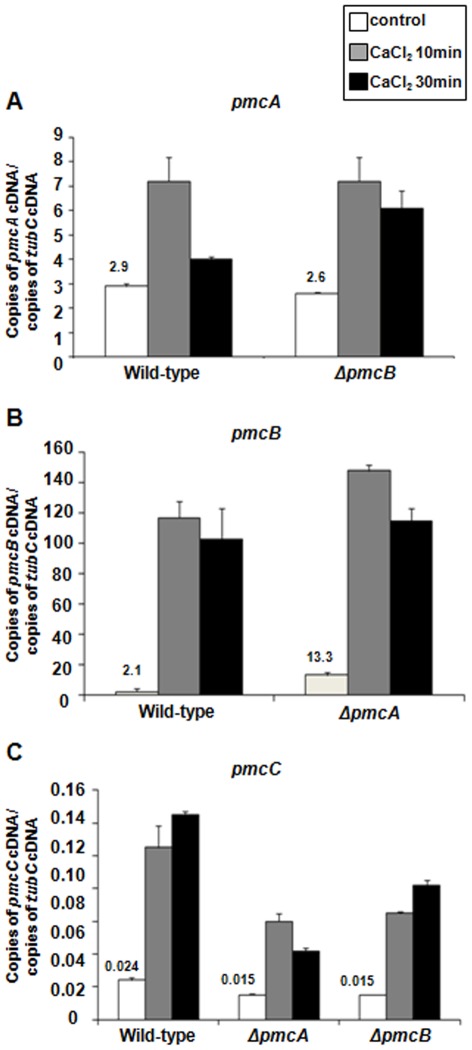
We also compared the absolute levels of mRNA abundance among pmcA, -B, and –C when the A. fumigatus wild-type, ΔpmcA and ΔpmcB mutant strains were exposed to 200 mM CaCl2 (Figure 4). Upon exposure of wild-type A. fumigatus to calcium, pmcB mRNA levels were higher than pmcA and pmcC, while pmcA levels were higher than pmcC (Figures 4A–C). The number of normalized pmcA and pmcC transcripts in the ΔpmcB and ΔpmcA mutant strains, respectively, were not different from the wild-type strain (Figures 4A and B), suggesting the absence of either pmcB or pmcA does not considerably affect the mRNA abundance of pmcA and pmcC. However, before adding 200 mM CaCl2 there was approximately six times more pmcB transcripts in the ΔpmcA than in the wild-type strain (Figure 4B). Interestingly, the pmcC mRNA levels are reduced upon CaCl2 exposure in both ΔpmcB and ΔpmcA mutant strains. These results suggest that there is compensation in the mRNA accumulation of pmcB in the ΔpmcA mutant strain and pmcC mRNA accumulation is dependent on pmcA and pmcB. Upon calcium exposure, down-regulation or overexpression of pmcC had no effect on pmcA or pmcB mRNA accumulation (data nor shown).
Figure 4. The pmcA-C genes have increased mRNA abundance when exposed to calcium.
The absolute quantitation of pmcA, pmcB, and pmcC and tubulin gene expression was determined by a standard curve (i.e., CT –values plotted against a logarithm of the DNA copy number). The results are the means (± standard deviation) of four biological replicates. (A–C) The mRNA abundance of pmcA-C in the wild-type, ΔpmcA, and ΔpmcB.
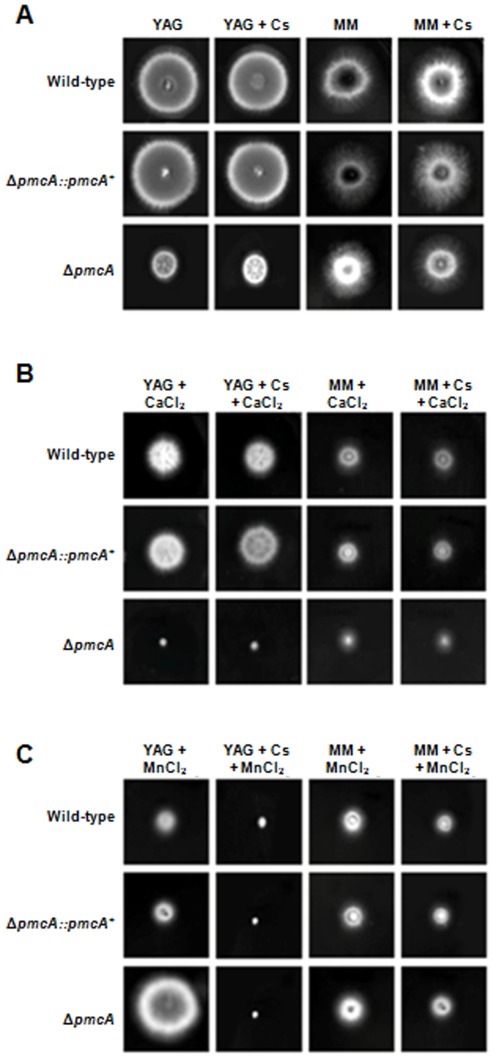
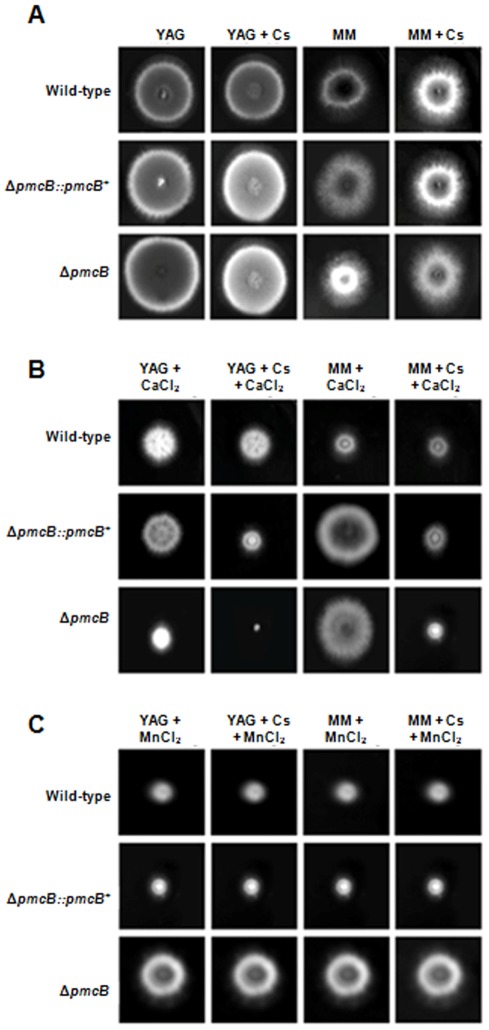
Next, we characterized the phenotype of ΔpmcA and ΔpmcB by growing these strains in different culture media in the presence and absence of cyclosporin A (CsA). This immunosuppressive drug inhibits calcineurin signaling by forming a complex with the immunophilin cyclophilin which then inhibits calcineurin [24]. In addition, since the ΔcrzA mutant is also sensitive to MnCl2 (Soriani et al., 2008), we decided to investigate a possible influence of pmcA-B on this phenotype. Curiously, the ΔpmcA mutant strain demonstrated different behavior in complete (YAG) and minimal media (MM) (Figure 5A). It showed reduced radial growth rate in complete medium when compared to the wild-type strain, but this reduction in growth was not suppressed by cyclosporin 25 ng/ml (Figures 5A). The ΔpmcA mutant strain was sensitive to CaCl2 500 mM and showed increased sensitivity in YAG and MM, compared to both the wild-type and other mutant strains, when cyclosporin 25 ng/ml was added (Figure 5B). The ΔpmcA mutant strain was resistant to MnCl2 25 mM in both YAG and MM media, however cyclosporin suppressed ΔpmcA resistance in YAG and wild-type sensitivity in MM (Figure 5C). The ΔpmcB mutant strain had about the same radial diameter than the wild-type strain in both MM and YAG media (Figure 6A), but it was much more sensitive to CaCl2 in YAG and showed increased sensitivity when grown in the presence of cyclosporin (Figure 6B). However, in MM+500 mM CaCl2 the ΔpmcB mutant strain has the same radial diameter as the wild-type strain (Figure 6B). In addition, the ΔpmcB mutant strain was more resistant to YAG+25 mM MnCl2 than the wild-type strain (Figure 6C), but this resistance was suppressed in the presence of cyclosporin (Figure 6C). The same growth was observed for both wild-type and ΔpmcB when grown in MM+25 mM MnCl2 (Figure 6C). Both the ΔpmcA::pmcA + and ΔpmcB::pmcB + showed the same phenotype as the wild-type strain, strongly indicating that the null phenotypes observed for both genes were only due to the introduction of these mutations in the corresponding strains (Figures 5 and 6).
Figure 5. Growth phenotypes of the ΔpmcA mutant strain.
The A. fumigatus wild-type, ΔpmcA::pmcA + and ΔpmcA mutant strains were grown for 72 hours at 37°C in (A)YAG, YAG+25 ng/ml cyclosporin (Cs), MM, or MM+25 ng/ml Cs; (B) YAG+500 mM CaCl2, YAG+25 ng/ml Cs+500 mM CaCl2, MM+500 mM CaCl2, or MM+25 ng/ml Cs+500 mM CaCl2; (C) YAG+25 mM MnCl2, YAG+25 ng/ml Cs+25 mM MnCl2, MM+25 mM MnCl2, or MM+25 ng/ml Cs+25 mM MnCl2.
Figure 6. Growth phenotypes of the ΔpmcB mutant strain.
The A. fumigatus wild-type, ΔpmcB::pmcB and ΔpmcB mutant strains were grown for 72 hours at 37°C in (A)YAG, YAG+25 ng/ml cyclosporin (Cs), MM, or MM+25 ng/ml Cs; (B) YAG+500 mM CaCl2, YAG+25 ng/ml Cs+500 mM CaCl2, MM+500 mM CaCl2, or MM+25 ng/ml Cs+500 mM CaCl2; (C) YAG+25 mM MnCl2, YAG+25 ng/ml Cs+25 mM MnCl2, MM+25 mM MnCl2, or MM+25 ng/ml Cs+25 mM MnCl2.
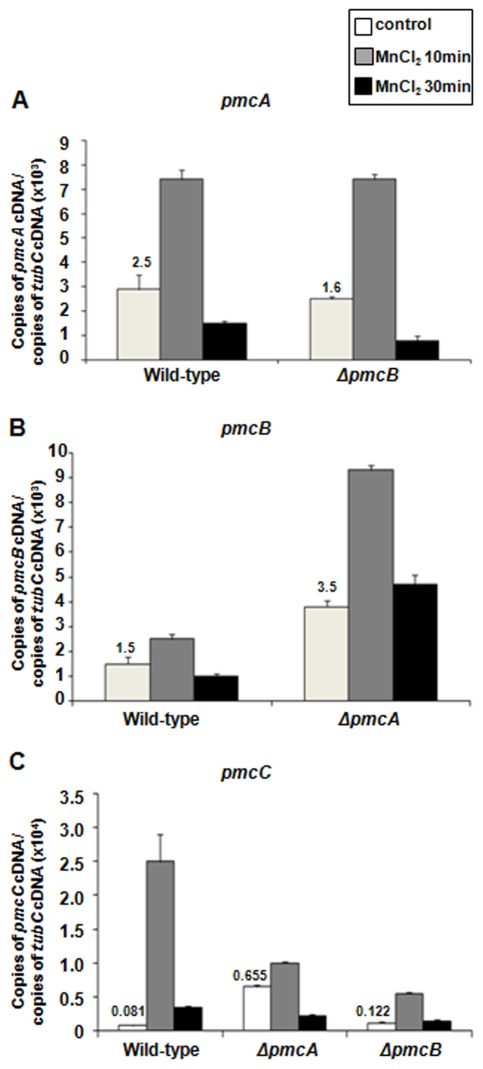
Since the ΔpmcA and ΔpmcB strains were calcium-sensitive but manganese-resistant, we decide to investigate the mRNA abundance of each pmc gene when the wild-type, ΔpmcA and ΔpmcB mutant strains were exposed to a short pulse of MnCl2 (Figure 7). All three genes showed increased mRNA abundance in the presence of MnCl2 (pmcA and pmcB have 2.5- and 2.0-fold more transcripts after 10 minutes; Figures 7A and B), however the highest induction was observed for pmcC that showed a 30- and 3.7-fold increase in transcripts after 10 and 30 minutes, respectively (Figure 7C). Nevertheless, like observed for calcium induction, the absolute mRNA levels of pmcC are the lowest among all the three genes (Figure 7). The pmcA mRNA levels in the ΔpmcB mutant strain exposed to MnCl2 were about the same as the wild-type strain (Figure 7A). When the ΔpmcA mutant strain was exposed to MnCl2, the mRNA levels of pmcB were 2.5-fold higher than the wild-type strain after 10 minutes exposure. Interestingly, the pmcB mRNA levels in this mutant without any MnCl2 exposure (i.e., the control before exposure) were 2.3-fold higher than the wild-type strain (Figure 7B). Finally, there was a decrease in the pmcC mRNA levels after ΔpmcA and ΔpmcB mutant strains were exposed to MnCl2 (Figure 7C). Down-regulation or overexpression of pmcC had no effect on pmcA or pmcB mRNA accumulation (data nor shown).
Figure 7. The pmcA-C genes have increased mRNA abundance when exposed to manganese.
The absolute quantitation of pmcA, pmcB, and pmcC and tubulin gene expression was determined by a standard curve (i.e., CT –values plotted against a logarithm of the DNA copy number). The results are the means (± standard deviation) of four biological replicates. (A–C) The mRNA abundance of pmcA-C in the wild-type, ΔpmcA, and ΔpmcB.
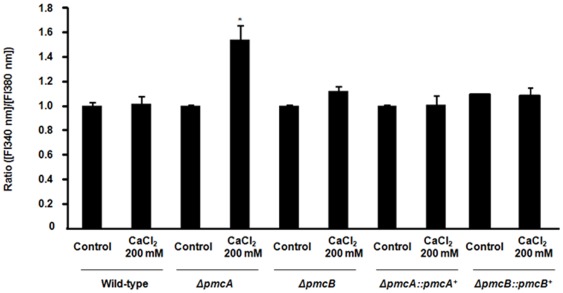
Finally, we evaluated the relative concentration of free calcium in the A. fumigatus wild-type, ΔpmcA, ΔpmcB, ΔpmcA::pmcA +, and ΔpmcB::pmcB + strains by using Fura-2-AM, a highly sensitive dye for rapid measurement of calcium flux in cells (www.invitrogen.com). Fura-2-Am is a fluorescent calcium indicator that can passively diffuse across cell membranes and when inside the cell, the esters are cleaved by intracellular esterases to yield cell-impermeant fluorescent indicator. Upon binding Ca+2, Fura-2 exhibits an absorption shift from 380 to 340 nm of excitation. Thus, the relative Ca+2 concentration was evaluated based on the fluorescence ratio after dual-wavelength excitation. Upon calcium exposure, the ΔpmcA mutant strain had an increased relative level of intracellular calcium concentration compared to the same strain in the absence of calcium (Figure 8). This difference is not observed for the wild-type, ΔpmcB and complemented strains, and alcA::pmcC strain (data not shown).
Figure 8. The ΔpmcA mutant strain has increased accumulation of calcium in the cytoplasm.
The relative levels of intracellular calcium in the wild-type, ΔpmcA and ΔpmcB mutant strains were determined using the calcium-sensitive dye Fura-2-AM. The relative Ca+2 concentration was determined based on the fluorescence ratio after dual-wavelength excitation (fluorescent intensity at 340 nm [FI340 nm]/[FI380 nm]. Data shown are means of three repetitions ± standard deviations. Statistical analysis was performed by using either One-Way Anova with Newman-Keuls post-tests or Tukey's multiple comparison tests. *p<0.005.
We have not observed any differential susceptibility of these mutants to antifungal agents, such as amphotericin, azoles, and caspofungin, in E-tests (data not shown). These results indicate pmcA and pmcB are involved in calcium and manganese metabolism in A. fumigatus, and also suggest pmcA is the major transporter responsible for removing calcium from the cytoplasm.
Expression of the pmcA-C genes in murine-infecting A. fumigatus wild-type, ΔcalA and ΔcrzA mutant strains
Previously, we generated by RNA amplification multiple gene expression profiles via minute samplings of A. fumigatus germlings during the initiation of murine infection [25], [26]. This enabled us to identify genes preferentially expressed during adaptation to the mammalian host niche. Here, we took advantage of the establishment of this technical platform to characterize genes that have in vivo decreased or increased mRNA abundance in the ΔcalA and ΔczA mutant strains when compared to the wild-type strain. We firstly characterized the time course of hyphal development in the sequenced clinical isolate Af293, ΔcalA and ΔcrzA mutant strains by histopathological examination of infected neutropenic murine lung tissues (Supplementary Figure S4). Lung sections collected and formalin-fixed at 4, 10 and 14 hours post-infection contained numerous A. fumigatus wild-type, ΔcalA and ΔcrzA spores in close association with murine epithelium in the bronchioles and alveoli (Supplementary Figure S4, upper panels). At 12–14 hours post-infection, 80% of A. fumigatus conidia from the three strains had undergone comparable germination and primary hyphal production. Bronchoalveolar lavage was performed immediately using pre-warmed sterile saline and samples (BALFs) were snap frozen prior to RNA extraction and amplification. Within infection groups BALFs were pooled prior to RNA extraction and mRNA amplification. Total RNA extracted from these cultures was used to amplify fluorescent-labeled cDNAs for real-time PCR experiments. We designed Lux fluorescent probes and used real-time RT-PCR analysis to quantify the pmcA, pmcB, and pmcC mRNA abundance in the ΔcalA and ΔcrzA germlings after bronchoalveolar lavage at 4 and 14 hours growth and compared this with their expression in the wild-type strain grown during the same time points (reference treatment) (Table 1). All these three genes showed, to different extents, decreased mRNA abundance during the initiation of murine infection by the A. fumigatus ΔcalA and ΔcrzA mutant strains relative to the wild-type strain. Thus, it seems that in vivo pmcA-C mRNA accumulation is dependent on CalA and crzA.
Table 1. Real-time RT-PCR for pmcA-C genes from the in vivo microarray.
| Gene* | Wild-type4 hs | Wild-type14 hs | ΔCalA4 hs | ΔCalA14 hs | ΔcrzA4 hs | ΔcrzA14 hs |
| pmcA (Afu1g10880) | 0.38±0.01 | 0.22±0.00 | 0.20±0.00 | 0.09±0.00 | 0.17±0.01 | 0.11±0.00 |
| pmcB (Afu3g10690) | 1.52±0.02 | 3.46±0.23 | 0.23±0.00 | 0.32±0.06 | 0.35±0.08 | 0.56±0.00 |
| pmcC (Afu7g01030) | 0.01±0.00 | 0.01±0.00 | 0.00±0.00 | 0.00±0.00 | 0.00±0.00 | 0.00±0.00 |
The mRNA abundance of A. fumigatus pmcA-C genes during growth in lung alveoli. Real-time RT-PCR was used to quantify mRNA abundance. The measured quantity of mRNA for a specific gene in each of the treated samples was normalized using the CT values obtained for the β-tubulin mRNA amplifications run on the same plate. The relative quantitation of a specific gene and β-tubulin gene expression was determined by a standard curve (i.e., CT –values plotted against a logarithm of the DNA copy number). The results of four sets of experiments were combined for each determination; means ± standard deviation are shown. The values represent the cDNA concentration of a specific gene divided by the β-tubulin cDNA concentration.
The A. fumigatus ΔpmcA mutant strain is avirulent in low dose murine infection
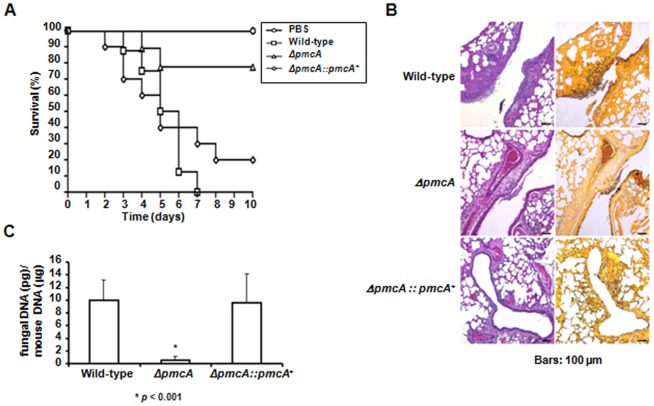
To assess the role of PmcA-B in pathogenicity we tested the A. fumigatus ΔpmcA-B mutant strains in a neutropenic murine model of invasive pulmonary aspergillosis, comparing virulence of the A. fumigatus ΔpmcA-B mutant strains (n = 10 for each mutant) to that of the wild-type (n = 10) (Figure 9A and Supplementary Figure S5). While infection with the wild-type strain resulted in a mortality rate of over 100% at 6 days post-infection, infection with the pmcA deletion strain resulted in a significantly reduced mortality rate of approximately 20% after 10 days post-infection (p<0.005). The pmcB mutant showed virulence comparable to the wild-type strain (Supplementary Figure S5). Since the comparison between ΔpmcA infected group and the non-infected group (PBS) showed to be statistically non-significant (p = 0.1451), we can consider this strain avirulent. To directly link the observed attenuated virulence of the ΔpmcA mutant with the replacement of the ΔpmcA locus we tested an independent strain resulting from single ectopic reintegration of the wild-type ΔpmcA locus (Supplementary Figure S6) and with the complementation strain full virulence was restored (Figure 9A). To further understand the basis of attenuated virulence in the ΔpmcA background we made histopathological examinations of infected tissues at early time points in infection, aiming to identify differences in growth rate, tissue invasion and inflammatory responses between the two strains. At 72 hours post-infection the lungs of mice infected with the wild-type isolate contained multiple foci of invasive hyphal growth, manifesting as both penetration of the pulmonary epithelium in major airways (Figure 9B) and pockets of branched invading mycelia originating from the alveoli (Figure 9B). In contrast, infection resulting from ΔpmcA inoculations was typified by contained inflammatory infiltrates in bronchioles (Figure 9B) some of which contained fungal elements in the form of poorly germinated or ungerminated spores. Fungal burden data as measured by real-time PCR showed that the ΔpmcA mutant strain did not grow within the lungs as well as the wild-type and the complemented ΔpmcA strains (Figure 9C, p<0.0001). Taken together, these data strongly indicate that PmcA plays a role in A. fumigatus virulence.
Figure 9. A. fumigatus pmcA contributes to virulence in neutropenic mice.
(A) Comparative analysis of wild-type, ΔpmcA and ΔpmcA::pmcA + strains in a neutropenic murine model of pulmonary aspergillosis. A group of 10 mice per strain was infected intranasally with a 20 µl suspension of conidiospores at a dose of 5.0×104. (B) Histological analysis of infected murine lung were performed 72 hours after infection with the wild-type strain reveals invasion of the murine lung epithelium (C) Fungal burden was determined 48 hours post-infection by real-time RT-PCR based on 18S rRNA gene of A. fumigatus and an intronic region of the mouse GAPDH gene. Fungal and mouse DNA quantities were obtained from the Ct values from an appropriate standard curve. Fungal burden was determined through the ratio between ng of fungal DNA and µg of mouse DNA quantities. The results are the means (± standard deviation) of five lungs for each treatment.
Discussion
We have been actively looking for additional components of the Ca+2-calcineurin pathway [14], [17], [27]. One of these components, the transcription factor CrzA induces the expression of various cation transporters that act at the plasma membrane or on other membranous organelles [14], [17]. Very little is known about calcium transport and calcium homeostasis in filamentous fungi. Most of our knowledge about calcium homeostasis in fungi is derived from S. cerevisiae, where more than 95% of cellular calcium is sequestered in the vacuole [28], [29], [30]. In S. cerevisiae PMC1, PMR1, and VCX1 encode a vacuolar Ca2+ ATPase involved in depleting cytosol of Ca2+ ions, a high affinity Ca2+/Mn2+ P-type ATPase required for Ca2+ and Mn2+ transport into the Golgi, and a vacuolar membrane antiporter with Ca2+/H+ and K+/H+ exchange activity, involved in the control of cytosolic Ca2+ and K+ concentrations, respectively [19], [22], [31]. S. cerevisiae PMC1 knock-out mutants sequester Ca+2 into the vacuole at 20% of the wild-type levels and fail to grow in media containing high levels of Ca+2 [19]. Mutations in the calcineurin A or B subunits or the addition of FK506 or cyclosporin A restored growth of pmc1 mutants in media with high Ca+2 concentrations [20], [21]. In Neurospora crassa it was reported that active transport across the plasma membrane is important for keeping low levels of cytosolic calcium [32], [33]. In addition, in this species the vacuole is important for regulating the intracellular calcium levels [32], [34], [35]. In N. crassa, there are two PMC1 homologues, NCA-2 and NCA-3 [36]. The NCA-2 fused with GFP is located in the plasma membrane as well as in vacuolar membranes in this organism [37], suggesting NCA-2 functions to pump calcium out of the cell. The Δnca-3 strain showed comparable levels of calcium sensitivity to the wild-type strain; in contrast, Δnca-2 showed significant inhibition of growth at 50 mM CaCl2 and accumulates 10-fold more intracellular calcium than the wild-type strain [36]. In A. nidulans, two null mutants constructed for the PMC1 homologues, pmcA and pmcB, displayed low-sensitivity to 700 mM CaCl2 concentrations [38]. However, the double A nidulans ΔpmcA ΔpmcB mutant has increased calcium-sensitivity suggesting these two genes are genetically interacting [38].
Rispail et al. [39] have proposed that A. fumigatus has three PMC1, two PMR1, and four VCX1 homologues. Here, we have concentrated our attention on three genes encoding PMC1 calcium transporter homologues that have their mRNA levels dependent on CalA-CrzA [14], [17], generated mutants of them, and studied their phenotypes and virulence. Although A. fumigatus pmcA-C genes are involved in calcium metabolism, this work did not provide a full characterization of their function. We do not know their sub-cellular localization and how they affect sub-cellular calcium abundance. We were not able to knock-out pmcC and subsequently demonstrated that pmcC is an essential A. fumigatus gene by constructing a conditional pmcC mutant. The pmcC downregulation causes growth inhibition and its overexpression can produce a physiological imbalance, as the mutant strain also has reduced growth. We were able to demonstrate that CrzA can control the pmcA-C mRNA expression by binding directly to their promoter regions. Crz1p has a C2H2 zinc finger motif that binds to CDRE in the promoters of genes that are regulated by calcineurin and calcium (Stathopoulos and Cyert, 1997). Yoshimoto et al. [40] have identified the S. cerevisiae Crz1p-binding site as 5′-GNGGC(G/T)CA-3′ by in vitro site selection. Recently, Hagiwara et al. [41] identified and characterized the A. nidulans AncrzA gene. They performed an in silico analysis by also using MEME of the possible presence of a CDRE-like consensus motif in the promoter regions of 25 AnCrzA-dependent genes. By analyzing their promoter regions, 5′-G[T/G]GGC[T/A]G[T/G]G-3′ was presumed to be the consensus sequence for the A. nidulans AnCrzA-dependent genes. By using a combination of MEME analysis and the A. nidulans CDRE consensus as a guide, we were able to identify 28 A. fumigatus genes that were repressed in ΔcrzA mutant strain upon CaCl2 exposure (Soriani et al., 2010), with 5′-GT[T/G]G[G/C][T/A]GA[G/T]-3′ as the CDRE-consensus sequence for A. fumigatus AfCrzA-dependent genes. Here, we demonstrated that CrzA can bind directly to 300-bp upstream regions from pmcA-C genes. In two of these genes, pmcA and pmcC, we were able to identify putative CDRE motifs and demonstrated that they can completely inhibited the complexes formed with pmcA and pmcC DNA fragments. These results strongly suggest these CDRE motifs are functional and this is probably the first demonstration of CDRE functionality in a human pathogenic fungus.
Cyclosporin was able to modulate the sensitivity or resistance of these mutants to either calcium or manganese chloride, once more supporting the interaction between calcineurin and the function of these transporters. In addition, we showed wild-type levels of susceptibility to amphotericin B, voriconazole, posoconazole, itraconazole, and caspofungin (E-test assays) and that there were no defects in cell wall integrity (data not shown). We also observed that the complete and minimal culture media affected the susceptibility of the ΔpmcA and ΔpmcB mutant strains to calcium and manganese chloride. The defined macronutrients composition of MM medium could explain the differences in growth of the ΔpmcA and ΔpmcB mutant strains in YAG and MM media. The MM is composed of glucose, trace elements, and macronutrients (salt solution). The salt solution is composed of sodium nitrate, potassium chloride, potassium phosphate, and magnesium sulphate. When the wild-type, ΔpmcA, and ΔpmcB strains are grown in MM supplemented only with a single one of these macronutrients, there is a reduction in radial growth for all strains, except for MM+MgSO4 that showed about the same radial growth as in MM (data not shown). The most likely reasons for this outcome are either the mechanism of action of these transporters depends on other cations, such as sodium, or there is some cross-talk with the mechanisms for ion detoxification. Recently, Spielvogel et al. [42] have shown that SltA, a transcription factor important for cation adaptation and homeostasis acts positively on the transcription of the Ena1p-like Na+ pump gene enaA and negatively on the transcription of the putative vacuolar Ca+2/H+ exchanger gene vcxA (A. fumigatus homologue is Afu1g04270). Interestingly, the negative regulation of vcxA by SltA is opposed by its transcriptional activation by CrzA [42].
A. fumigatus pmcA-C genes have decreased mRNA abundance into the alveoli in the ΔcalA and ΔcrzA mutant strains. Accordingly, when A. fumigatus is exposed in vitro to calcium chloride, there is a decrease in pmcA-C mRNA abundance in both mutants. When we compare the absolute pmcA-C mRNA abundance levels in the wild-type strain grown in mouse alveoli, we observed that pmcB has about five to ten times higher levels than pmcA, while pmcC has very low levels of mRNA abundance (1,000 to 3,000 times lower than pmcB). Consistently, the same mRNA abundance is observed when A. fumigatus is exposed in vitro to calcium chloride. Interestingly, there is an increase in the pmcB mRNA levels in the ΔpmcA mutant strain when this strain is not exposed to CaCl2, suggesting a compensation for the pmcA absence. An intriguing observation from our work is the fact that pmcC has very low absolute levels of mRNA accumulation in all conditions tested in this work, but it is an essential gene. This is confirmed by a weak CrzA binding to pmcC promoter. It is possible that PmcC specific activity is very high and this will compensate its low mRNA levels. It is also possible that PmcC has other functions that were not identified in this work and are essential for cell metabolism. Interestingly, both ΔpmcA and ΔpmcB mutants are more resistant to MnCl2 than the wild-type strain and had reduced pmcC mRNA accumulation when exposed to either CaCl2 or MnCl2. These results suggest pmcC mRNA levels are dependent on pmcA and pmcB, when A. fumigatus is exposed either to calcium or manganese. However, this effect is more notable in the presence of manganese.
The ΔpmcA mutant is avirulent in a neutropenic murine model of invasive pulmonary aspergillosis. The reduced virulence of the ΔpmcA could be due to an excess of calcium in the cytoplasm that could not be removed due to the lack of pmcA, thus potentially affecting several functions such as secretion, cell wall composition and the activation of pathways necessary for infection. Interestingly, we did not observe attenuated virulence for ΔpmcB, suggesting that the different PMC1 paralogues have different functions during pathogenicity. This is the first demonstration of the involvement of a calcium transporter in A. fumigatus virulence. Previously, Pinchai et al. [16] have shown that A.fumigatus ΔpmrA has several defects related to growth, cationic tolerance, and increased beta-glucan and chitin content, but in spite of all these abnormal phenotypes the mutant strain remained virulent.
In conclusion, we have shown that PmcA is required for full virulence in animal infection. In addition, that PmcA acts in the A. fumigatus Ca+2-calcineurin signaling pathway and influences the relative intracellular calcium concentration. Further studies are necessary to address the sub-cellular location of PmcA, -B, and –C, and how PmcA contributes to the pathogenesis of aspergillosis.
Materials and Methods
Ethics statement
The principles that guide our studies are based on the Declaration of Animal Rights ratified by the UNESCO in January 27, 1978 in its articles 8th and 14th. All protocols used in this study were approved by the local ethics committee for animal experiments from the Campus of Ribeirão Preto from Universidade de Sao Paulo (Permit Number: 08.1.1277.53.6; studies on the interaction of Aspergillus fumigatus with animals). All animals used in this study were housed in groups of five in individually ventilated cages and were cared for in strict accordance to the principles outlined in the by the Brazilian College of Animal Experimentation (Princípios Éticos na Experimentação Animal - Colégio Brasileiro de Experimentação Animal, COBEA) and Guiding Principles for Research Involving Animals and Human Beings, American Physiological Society. All efforts were made to minimize suffering. Animals were clinically monitored at least twice daily by a veterinarian and humanely sacrificed if moribund (defined by lethargy, dyspnoea, hypothermia and weight loss).
Strains and culture conditions
The A. fumigatus strains used in this study are CEA17 (pyrG−), Af293 (wild-type), ΔcalA and ΔcrzA (Soriani et al., 2008), CEA17-80 (as the wild-type in all the experiments), ΔpmcA (ΔpmcA::pyrG), ΔpmcB (ΔpmcB::pyrG), ΔvcxA (ΔvcxA::pyrG). ΔpmcA::pmcA + and alcA::pmcC. The media used were of two basic types, i.e. complete and minimal. The complete media comprised the following three variants: YAG (2% w/v glucose, 0.5% w/v yeast extract, 2% w/v agar, trace elements), YUU [YAG supplemented with 1.2 g l-1 (each) of uracil and uridine], and liquid YG or YG + UU medium with the same composition (but without agar). A modified minimal medium (MM: 1% glucose, original high nitrate salts, trace elements, 2% agar, pH 6.5) was also used. Expression of pmcC gene, under the control of alcA promoter, was regulated by carbon source: repression on glucose 4% w/v, derepression on glycerol, and induction on threonine. Therefore, MM + Glycerol and MM + Threonine were identical to MM, except that glycerol (2% v/v) and/or threonine (100 mM) were added, respectively, in place of glucose as the sole carbon source. Trace elements, vitamins, and nitrate salts were included as described by [43]. Strains were grown at 37°C unless indicated otherwise. Additionally, 10% fetal bovine serum (Gibco) was used as a medium.
Construction of the A. fumigatus mutants
A gene replacement cassette was constructed by “in vivo” recombination in S. cerevisiae as previously described [44]. Briefly, approximately 2.0 kb regions on either side of each ORF were selected for primer design. For construction, the primers were named as 5F and 5R, were used to amplify the 5′-UTR flanking region of the targeted ORF. Likewise, the primers 3F and 3R were used to amplify the 3′-UTR ORF flanking region, and the primers 5F and 3R also contains a short homologue sequence to the MCS of the plasmid pRS426. Both fragments, 5- and 3-UTR, were PCR-amplified from A. fumigatus genomic DNA (gDNA). The pyrG used in the A. fumigatus cassette for generating the mutant strains were used as marker for prototrophy. Deletion cassette generation was achieved by transforming each fragment along with the plasmid pRS426 BamHI/EcoRI cut in the in S. cerevisiae strain SC94721 by the lithium acetate method [45]. The DNA of the yeast transformants was extracted by the method described by Goldman et al. [46], dialysed and transformed by electroporation in Escherichia coli strain DH10B to rescue the pRS426 plasmid harboring the cassette. The cassette was PCR-amplified from these plasmids and used for A. fumigatus transformation. Southern blot analyses were used throughout of the manuscript to demonstrate that the transformation cassettes had integrated homologously at the targeted A. fumigatus loci. For the construction of the alcA::pmcC strain, 1000 bp of the pmcC encoding region was cloned downstream to the alcA promoter into the pMCB17apx vector. This construction was further transformed in A. fumigatus to replace the endogenous pmcC promoter yielding the strain alcA::pmcC. The ΔpmcA mutant strain was complemented by co-transforming a pmcA + DNA fragment (approximately 1 kb from each 5′ and 3′-flanking regions plus the ORF) together with the pHATα vector [47] and selecting for hygromycin resistance in MM plates with 150 µg/ml of hygromycin B.
RNA extraction and real-time PCR reactions
After treatment conditions, mycelia were harvested by filtration, washed twice with H2O and immediately frozen in liquid nitrogen. For total RNA isolation, the germlings were disrupted by grinding in liquid nitrogen with pestle and mortar. Total RNA was extracted with Trizol reagent (Invitrogen, USA). Ten micrograms of RNA from each treatment was then fractionated in 2.2 M formaldehyde, 1.2% w/v agarose gel, stained with ethidium bromide, and then visualized under UV light. The presence of intact 25S and 18S ribosomal RNA bands was used to assess the integrity of the RNA. RNasefree DNase I treatment, for the real-time PCR experiments, was carried out as previously described [48]. Twenty micrograms of total RNA was treated with DNase, purified using a RNAeasy kit (Qiagen) and cDNA was generated using the SuperScript III First Strand Synthesis system (Invitrogen) with oligo(dT) primers, according to the manufacturer's protocol.
All the PCR reactions were performed using an ABI 7500 Fast Real-Time PCR System (Applied Biosystems, USA) and Taq-Man Universal PCR Master Mix kit (Applied Biosystems, USA). The reactions and calculations were performed according to Semighini et al. [48]. The primers and Lux™ fluorescent probes (Invitrogen, USA) used in this work are described in Supplementary Table S1.
Cloning the crzA gene into the pDEST15 vector
The Gateway Technology (Invitrogen) was used to construct, in Escherichia coli, the expression system consisting of the CRZA gene N-tagged to the GST gene. Briefly, the coding region of the exon 2 from CrzA was amplified from the cDNA sample by PCR using Platinum® Taq DNA Polymerase High Fidelity. (Invitrogen) and specific primers (CRZ-exon2-attB1-F 5′- GGGGACAAGTTGTACAAAAAAGCAGGCTTCGAAGGAGATAGAACCATGTCCCGCGGGCGTAGCAAG-3′ and CRZ-attB2-R 5′- GGGGACCACTTTGTACAAGAAAGCTGGGTCTCAATAGAAGTTACCGGCAGCAG-3′). Amplification was run for 30 cycles consisting of denaturation at 94°C for 1 min, primer annealing at 55°C for 1 min and primer extension at 68°C for 2 min. The PCR product carrying the attB sites was purified from agarose gel using the QIAquick PCR purification kit (Qiagen) and cloned into the pDONR201 plasmid (Invitrogen) using the BP Clonase. The BP clonase catalyze the in vitro recombination of PCR products or DNA segments from clones (containing attB sites) and a donor vector (containing attP sites) to generate entry clones. The entry clone pDONR201-CrzA was transformed into E. coli DH10B competent cells and selected for kanamicyn resistence. Entry clones were checked by sequencing using the ATT primers (ATT1F- 5′- TCGCGTTAACGCTAGCATGGATCTC-3′ and ATT2 R- 5′- GTAACATCAGAGATTTTGAGACAC-3′) and further used in LR reactions (Invitrogen) with the vector pDEST15 (an N-terminal GST fusion vector containing the T7 promoter) to generate the expression vector pDEST15-GST/CrzA.
Production and Purification of GST::CrzA
A. nidulans CrzA was expressed as a GST-fusion protein from the construct pDEST15-GST/CrzA in E. coli Rosetta™ (DE3) pLysS strain (Novagen). Cells harboring the plasmid construction were grown in 1 L of LB medium to an O.D.600 nm of 0.8 and protein expression was induced at 12°C, 180 rpm overnight with 0.4 mM IPTG final concentration. After induction, cells were harvested by centrifugation, suspended in phosphate-buffered saline solution (500 mM NaCl, 2.7 mM KCl, 100 mM Na2HPO4, 2 mM KH2PO4, 5% v/v glycerol, 0.5% NP-40, pH 7.4) containing 10 mM benzamidine, 0.5 mM EDTA and 2 mM of each DTT and PMSF and lysed by sonication (ten 30 sec pulses on ice) in a Vibra-Cell disrupter (Sonics®). Cell lysate was clarified at 23,000× g, 20 min, 4°C, and the recombinant protein was purified by affinity chromatography on a GSTrap FF column (GE HealthCare) according to manufacturer's instructions on an ÄKTA Prime purification system. Recombinant protein was eluted in a linear gradient of 20 mM glutathione in 50 mM Tris-HCl, 500 mM NaCl, 5% v/v glycerol, 2 mM DTT, pH 8.0 buffer. Chromatographic fractions were analyzed by SDS-PAGE followed by Coomassie Brilliant Blue staining [49] and fractions containing the purified protein were combined, concentrated and quantified using BSA as standard [50].
Electrophoretic Mobility Shift Assay
GST::CrzA recombinant protein was assayed in DNA-protein binding reactions using three 300 bp DNA fragments of the pmcA, pmcB and pmcC promoters as probes, containing the putative cis-regulatory calcineurin-dependent response elements (CDREs) for the transcription factor CrzA (Supplementary Table S1 and Figure S1). Binding reactions were carried out in 1×binding buffer (25 mM HEPES- KOH, pH 7.9, 20 mM KCl, 10% w/v glycerol, 1 mM DTT, 0.2 mM EDTA, pH 8.0, 0.5 mM PMSF, 12.5 mM benzamidine, 5 mg/mL of each antipain and pepstatin A) containing 2 µg poly(dI-dC).(dI-dC) as non-specific competitor and 1–2 µg of GST::CrzA recombinant protein, at room temperature for 10 min. After that, DNA probes (104 cpm) were added and the binding reactions were incubated at room temperature during 20 min prior to being loaded onto a native 5% polyacrylamide gel in 0.5× TBE buffer. Gels were run at 10 mA, 15°C, dried, and exposed to X-ray film. For competition assays, a molar excess of the specific DNA competitors were added prior to incubation with the radiolabeled probe.
DNA Probes and Specific Competitors for EMSA
Putative cis CrzA motifs were visually identified in the promoter regions of the genes pmcA, pmcB and pmcC by using the A. fumigatus CDRE consensus. To produce the pmcA probe, a 300 bp DNA fragment of the pmcA promoter was amplified from A. fumigatus genomic DNA by using the primers PMCA-5R and 5′-PMCA (Supplementary Table S1) in the presence of [α-32P]-dATP (3,000 Ci/mmol) and purified on 2% low-melting point agarose gel. pmcB and pmcC probes were prepared as above using the primer pairs PMCB-5R and 5′-PMCB (Supplementary Table S1), and PMCC-5R and 5′-PMCC (Supplementary Table S1). The unlabeled 300 bp pmcA, pmcB and pmcC probes were used as specific DNA competitors which were quantified by measuring the absorbance at 260 nm and added to the binding reaction in a 30- to 50-fold molar excess, 10 min prior to the addition of the respective probes. DNA oligonucleotides containing the CDRE motifs identified in pmcA and pmcC probes were also used as specific competitors after annealing the complementary oligonucleotides pairs pmcA1/pmcA2 and pmcC1/pmcC2, respectively (Supplementary Table S1). The DNA oligonucleotides were quantified by measuring the absorbance at 260 nm and added to the reaction in 10–30 fold molar excess.
Mutated probes (mpmcA and mpmcC) were prepared by changing the element core sequences by site-directed mutagenesis in a two-step PCR. In the mpmcA probe the sequence 5′-CCCTGCCCC-3′ was changed to 5′-AAAGTAAAA-3′ by using the oligonucleotide pair mPMCA-F and mPMCA-R in the in the first reaction to amplify two fragments. The oligonucleotide pair PMCA-5R and 5′-PMCA (Supplementary Table S1) was used in a second reaction to amplify the whole DNA fragment containing the mutation. In the mpmcC probe the sequence 5′-CACAGCCAC-3′ was changed to 5′-ACACTAACA-3′ by using the oligonucleotide pair mPMCC-F and mPMCC-R in the in the first reaction. The oligonucleotide pair PMCC-5R and 5′-PMCC (Supplementary Table S1) was used in a second reaction to amplify the whole DNA fragment containing the mutation. For EMSA, both mutated fragments were used as templates in PCR amplifications in the presence of [α-32P]-dATP (3,000 Ci/mmol) and purified on 2% low-melting point agarose gel.
Determination of the relative levels of intracellular calcium concentration
To investigate the relative intracellular free calcium concentration we used the Fura-2 acetoxymethyl ester (Fura-2-AM; Invitrogen). Briefly, 107 conidia of each wild-type, ΔpmcA, ΔpmcA::pmcA+, ΔpmcB, and ΔpmcB::pmcB+ were incubated in YG medium for 8 hours with shaking at 37°C. Then, each strain was either treated with 500 mM CaCl2, or not, in fresh YG medium for 30 minutes. After incubation the cells were washed three times with PBS and loaded with 10 µM Fura-2-AM for 30 min at 37°C. After washing, Fura-2 fluorescence was measured by alternating the excitation wavelengths at 340 and 380 nm with an emission wavelength fixed at 505 nm. The relative intracellular calcium concentration is expressed as the ratio between fluorescence intensities with excitation wavelengths at 340 and 380 nm. All data presented are representative of three independent experiments.
Murine model of pulmonary aspergillosis
Outbred female mice (BALB/c strain, 20–22 g) were housed in individually vented cages, containing 5 animals. Mice were immunosuppressed with cyclophosphamide at 150 mg/kg of body weight, administered intraperitoneally on days −4, −1 and 2, and hydrocortisonacetate was injected subcutaneously at 200 mg/kg on day −3, modified from [51]. A. fumigatus spores for inoculation were grown on Aspergillus complete medium for 2 days prior to infection. Conidia were freshly harvested using sterile PBS and filtered through Miracloth (Calbiochem). Conidial suspensions were spun for 5 min at 3000 g, washed three times with sterile PBS, counted using a hemocytometer and re-suspended at a concentration of 2.5×106 conidia/ml. Viable counts from administered inocula were determined following serial dilution by plating on Aspergillus complete medium and grown at 37°C. Mice were anaesthetized by halothane inhalation and infected by intranasal instillation of 5.0×104 conidia in 20 µl of PBS. As negative control, a group of 5 mice received only PBS intranasally. Mice were weighed every 24 h from the day of infection and visually inspected twice daily. In the majority of cases the end-point for survival experimentation was when a 20% reduction in body weight measured from the day of infection and at this point the mice were sacrificed. Significance of comparative survival was calculated using Log Rank analysis in the Prism statistical analysis package. Additionally, at 3 days post infection, 2 mice per strain were sacrificed, from which the lungs were removed, fixed and processed for histological analysis.
Lung histopathology and fungal burden
After sacrifice, the lungs were removed and fixed for 24 h in 10% buffered formalin phosphate. Samples were washed in 70% alcohol several times, dehydrated in alcohols of increasing concentrations, diafanized in xylol and embedded in paraffin. For each sample, sequential 5 µm sections were collected on glass slides and the sections were stained with Gomori methenamine silver (GMS) or hematoxylin and eosin (HE) stain following standard protocols [24]. Briefly, sections were deparaffinized, oxidized with 4% chromic acid, stained with methenamine silver solution, and counter stained with picric acid or light green. For HE staining, sections were deparaffinized, stained first with hematoxylin and then stained with eosin. All stained slides were immediately washed, preserved with mounting medium and sealed with a cover glass. Microscopical analyses were done using an Axioplan 2 imaging microscope (Zeiss) at the stated magnifications under brightfield conditions.
To investigate fungal burden in murine lungs, mice were immunosuppressed with cyclophosphamide at 150 mg/kg of body weight administred intraperitoneally on days −4 and −1 and hydrocortisonacetat injected subcutaneously at 200 mg/kg on day −3. Five mice per group (wild-type, ΔpmcA, ΔpmcA::pmcA, and PBS control) were inoculated with 5×105 conidia/20 µl suspension intranasally. A higher inoculum, in comparison to the survival experiments, was used to increase fungal DNA detection. Animals were sacrificed 48 hours post infection, both lungs were harvested and immediately frozen in liquid nitrogen. A mortar and pestle were used to pulverize the samples (frozen in liquid nitrogen) and DNA was extracted by the Phenol/Chlroform method. DNA quantity and quality was assessed with a NanoDrop 2000 (Thermo Scientific). Around 200 ng of total DNA of each sample was used for quantitative Real-Time PCR reaction. A primer and a Lux™ probe (invitrogen) were used to amplify the 18S rRNA region of A. fumigatus (primer: 5′-CTTAAATAGCCCGGTCCGCATT-3′, probe: 5′-CATCACAGACCTGT TATTGCCG-3′) and an intronic region of mouse GAPDH (primer: 5′-CGAGGGACTTGGAGGACACAG-3′, probe: 5′-GGGCAAGGCTAAAGGTCAGCG-3′). Six-point standard curves were calculated using serial dilutions of gDNA from all A. fumigatus strains used here and non-infected mouse lung. Fungal and mouse DNA quantities were obtained from the Ct values from an appropriate standard curve. Fungal burden was determined via the ratio between ng of fungal and mouse DNA.
Bronchoalveolar lavages
To analyze gene expression of A. fumigatus strains during early pulmonary infection, Outbred female mice (BALB/c strain, 20–22 g) were housed in individually vented cages, containing 5 animals. Mice were immunosuppressed with cyclophosphamide (Genuxal, Baxter) at 150 mg/kg of body weight administered intraperitoneally on days −4 and −1 and hydrocortisone sodium succinate (Hidrosone, Cellofarm) was injected subcutaneously at 200 mg/kg on day −1. All mice received tetracycline hydrochloride 0.5 mg/L in drinking water, as prophylaxis against bacterial infection. A. fumigatus spores for inoculation were grown on Aspergillus complete solid medium (YAG) for 2 days prior to infection. Conidia were freshly harvested using sterile PBS and filtered through Miracloth (Calbiochem). Conidial suspensions were spun for 5 min at 4,000 rpm, washed three times with sterile PBS, counted using a hemocytometer and re-suspended at a concentration of 2.5×1010 conidia/ml. Five mice per group (wild-type, ΔcrzA and ΔcalA) were anesthetized by isoflurane (Isothane, Baxter) inhalation and infected by intranasal instillation of 109 conidia in 40 µl of PBS. Groups of infected mice were sacrificed and processed collectively at time points 4 and 12 hours post-infection. Bronchoalveolar lavage (BAL) was performed immediately after culling using three 0.5 ml aliquots of cold sterile PBS. To remove the mice cells from BALs, samples were spun down in microcentrifuge tubes, supernatants were removed, the samples were resuspended in 1 ml of sterile ultrapure water, centrifuged again and finally the pellets were snap frozen immediately using liquid nitrogen. To extract RNA, BAL samples from each strain (5 BALs per strain) were mixed with 1 ml Trizol LS Reagent (Invitrogen) and acid treated glass beads (425–600 µm, Sigma-Aldrich). Fungal cells were homogenized by 10 min vortexing, centrifuged at 12,000 rpm for 10 min, the upper phase was mixed with 200 µl chloroform, centrifuged again, the new upper phase was mixed with 500 µl isopropanol and incubated overnight at −80°C. After washing the pellet with 70% ethanol, RNA was dissolved in 20 µl DEPC water. Further RNA purification was carried out using RNeasy mini kit (Qiagen), following manufacturer's instructions. RNA concentration and integrity was monitored by NanoDrop® 2000 – Thermo Scientific (Uniscience). RNA amplification was done according to Agilent Low RNA Input Fluorescent Linear Amplification kit (Agilent Technologies). RNAse free DNAse treatment was carried out as previously described [48].
Supporting Information
(A) Clustal alignment of A. fumigatus PmcA, PmcB, and PmcC.
(DOCX)
(B) CrzA-binding regions of the pmcA-C genes (upstream the ATG start codon). Underline and in bold the putative CDRE-motif.
(DOCX)
Southern blot and PCR analyses for (A) ΔvcxA, (B) ΔpmcB, (C) ΔpmcA and (D) alcA::pmcC.
(PPT)
Histological analysis of alveolar lavages after infection with the A. fumigatus wild type, ΔcalA, and ΔcrzA strains. Germlings and host cells were detected by using Grocotts methenamine silver and haematoxylin and eosin staining, respectively. Bars, 100 µm.
(PPTX)
A. fumigatus ΔpmcB virulence studies in neutropenic mice. Comparative analysis of wild type and ΔpmcB strains in a neutropenic murine model of pulmonary aspergillosis. A group of 10 mice per strain was infected intranasally with 20 µl suspension of conidiospores at a dose of 2.0–5.0×104.
(PPTX)
(A) Growth phenotype of the ΔpmcA::pmcA+ strain grown on YAG+CaCl2 500 mM. (B) PCR of the pmcA open reading frame. (C) PCR of the pmcB open reading frame.
(PPT)
List of primers and probes used in this work.
(DOC)
Acknowledgments
We would like to thank Dr. Andréa Carla Quiapim and the LBMP- FFCLRP/USP for DNA sequencing. We also thank the reviewers for their suggestions and comments.
Footnotes
Competing Interests: Gustavo Henrique Goldman is currently serving as an editor for PLoS ONE. This does not alter the authors' adherence to all the PLoS ONE policies on sharing data and materials.
Funding: This research was supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Brazil. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Fox DS, Heitman J. Good fungi gone bad: the corruption of calcineurin. Bioessays. 2002;24:894–903. doi: 10.1002/bies.10157. [DOI] [PubMed] [Google Scholar]
- 2.Cyert MS. Calcineurin signaling in Saccharomyces cerevisiae: how yeast go crazy in response to stress. Biochem Biophys Res Commun. 2003;311:1143–1150. doi: 10.1016/s0006-291x(03)01552-3. [DOI] [PubMed] [Google Scholar]
- 3.Steinbach WJ, Reedy JL, Cramer RA, Perfect JR, Jr, Heitman J. Harnessing calcineurin as a novel anti-infective agent against invasive fungal infections. Nat Rev Microbiol. 2007;5:418–430. doi: 10.1038/nrmicro1680. [DOI] [PubMed] [Google Scholar]
- 4.Stie J, Fox D. Calcineurin regulation in fungi and beyond. Eukaryot Cell. 2008;7:177–186. doi: 10.1128/EC.00326-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stathopoulos-Gerontides A, Guo JJ, Cyert MS. Yeast calcineurin regulates nuclear localization of the Crz1p transcription factor through dephosphorylation. Genes Dev. 1999;13:798–803. doi: 10.1101/gad.13.7.798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karababa M, Valentino E, Pardini G, Coste AT, Bille J, et al. CRZ1, a target of the calcineurin pathway in Candida albicans. Mol Microbiol. 2006;59:1429–1451. doi: 10.1111/j.1365-2958.2005.05037.x. [DOI] [PubMed] [Google Scholar]
- 7.Stathopoulos AM, Cyert MS. Calcineurin acts through the CRZ1/TCN1-encoded transcription factor to regulate gene expression in yeast. Genes Dev. 1997;11:3432–3445. doi: 10.1101/gad.11.24.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zakrzewska A, Boorsma A, Brul S, Hellinngwerf KJ, Klis FM. Transcriptional response of Saccharomyces cerevisiae to the plasma membrane-perturbing compound chitosan. Eukaryot Cell. 2005;4:703–715. doi: 10.1128/EC.4.4.703-715.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matheos DP, Kingsbury TJ, Ahsan US, Cunningham KW. Tcn1p/Crz1p, a calcineurin-dependent transcription factor that differentially regulates gene expression in Saccharomyces cerevisiae. Genes Dev. 1997;11:3445–3458. doi: 10.1101/gad.11.24.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirayama S, Sugiura R, Lu Y, Maeda T, Kawagishi K, et al. Zinc finger protein Prz1 regulates Ca+2 but not Cl− homeostasis in fission yeast. J Biol Chem. 2003;20:18078–18084. doi: 10.1074/jbc.M212900200. [DOI] [PubMed] [Google Scholar]
- 11.Onyewu C, Wormley FL, Perfect JR, Jr, Heitman J. The calcineurin target, Crz1, functions in azole tolerance but is not required for virulence of Candida albicans. Infect Immun. 2004;72:7330–7333. doi: 10.1128/IAI.72.12.7330-7333.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santos M, de Larrinoa IF. Functional characterization of the Candida albicans CRZ1 gene encoding a calcineurin-regulated transcription factor. Curr Genet. 2005;48:88–100. doi: 10.1007/s00294-005-0003-8. [DOI] [PubMed] [Google Scholar]
- 13.Da Silva Ferreira ME, Heinekamp T, Hartl A, Brakhage AA, Semighini CP, et al. Functional characterization of the Aspergillus fumigatus calcineurin. Fungal Genet Biol. 2007;44:219–230. doi: 10.1016/j.fgb.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 14.Soriani FM, Malavazi I, da Silva Ferreira ME, Savoldi M, von Zeska Kress MR, et al. Functional characterization of the Aspergillus fumigatus CRZ1 homologue, CrzA. Mol Microbiol. 2008;67:1274–1291. doi: 10.1111/j.1365-2958.2008.06122.x. [DOI] [PubMed] [Google Scholar]
- 15.Cramer RAJ, Perfect BZ, Pinchai N, Park S, Perlin DS, et al. Calcineurin target crzA regulates conidial germination, hyphal growth, and pathogenesis of Aspergillus fumigatus. Eukaryot Cell. 2008;7:1085–1097. doi: 10.1128/EC.00086-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pinchai N, Juvvadi PR, Fortwendel JR, Perfect BZ, Rogg LE, et al. The Aspergillus fumigatus P-type Golgi apparatus Ca2+/Mn2+ ATPase PmrA is involved in cation homeostasis and cell wall integrity but is not essential for pathogenesis. Eukaryot Cell. 2010;9:472–6. doi: 10.1128/EC.00378-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soriani FM, Malavazi I, Savoldi M, Espeso E, Dinamarco TM, et al. Identification of possible targets of the Aspergillus fumigatus CRZ1 homologue, CrzA. BMC Microbiol. 2010;10:12. doi: 10.1186/1471-2180-10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pittman JK. Vacuolar Ca(2+) uptake. Cell Calcium. 2011;50:139–146. doi: 10.1016/j.ceca.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 19.Cunningham KW, Fink GR. Calcineurin-dependent growth control in Saccharomyces cerevisiae mutants lacking PMC1, a homolog of plasma membrane Ca2+ ATPases. J Cell Biol. 1994;124:351–63. doi: 10.1083/jcb.124.3.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cunningham KW. Acidic calcium stores of Saccharomyces cerevisiae. Cell Calcium. 2011;50:129–38. doi: 10.1016/j.ceca.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cunningham KW, Fink GR. Calcineurin inhibits VCX1-dependent H+/Ca2+ exchange and induces Ca2+ ATPases in Saccharomyces cerevisiae. . Mol Cell Biol. 1996;16:2226–2237. doi: 10.1128/mcb.16.5.2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olivero I, Corbacho I, Hernández LM. The ldb1 mutant of Saccharomyces cerevisiae is defective in Pmr1p, the yeast secretory pathway/Golgi Ca(2+)/Mn(2+)-ATPase. FEMS Microbiol Lett. 2003;219:137–42. doi: 10.1016/S0378-1097(03)00002-8. [DOI] [PubMed] [Google Scholar]
- 23.Flipphi M, Kocialkowska J, Felenbok B. Characteristics of physiological inducers of the ethanol utilization (alc) pathway in Aspergillus nidulans. Biochem J. 2002;15:25–31. doi: 10.1042/bj3640025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greenberger PA. Allergic bronchopulmonary aspergillosis. J Allergy Clin Immunol. 2002;110:685–92. doi: 10.1067/mai.2002.130179. [DOI] [PubMed] [Google Scholar]
- 25.McDonagh A, Fedorova ND, Crabtree J, Yu Y, Kim S, et al. Sub-telomere directed gene expression during initiation of invasive aspergillosis. PLoS Pathog. 2008;4:e1000154. doi: 10.1371/journal.ppat.1000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cairns T, Minuzzi F, Bignell E. The host-infecting fungal transcriptome. FEMS Microbiol Lett. 2010;307:1–11. doi: 10.1111/j.1574-6968.2010.01961.x. [DOI] [PubMed] [Google Scholar]
- 27.Malavazi I, da Silva Ferreira ME, Soriani FM, Dinamarco TM, Savoldi M, et al. Phenotypic analysis of genes whose mRNA accumulation is dependent on calcineurin in Aspergillus fumigatus. Fungal Genet Biol. 2009;46:791–802. doi: 10.1016/j.fgb.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 28.Dunn T, Gable K, Beeler T. Regulation of cellular Ca+2 by yeast vacuoles. J Biol Chem. 1994;269:7273–7278. [PubMed] [Google Scholar]
- 29.Eilam Y, Lavi H, Grossowicz N. Cytoplasmic Ca+2- homeostasis maintained by a vacuolar Ca+2 transport-system in the yeast Saccharomyces cerevisiae. J Gen Microbiol. 1985;131:623–629. doi: 10.1099/00221287-131-10-2555. [DOI] [PubMed] [Google Scholar]
- 30.Halachmi D, Eilam Y. Cytosolic and vacuolar Ca+2- concentrations in yeast cells measured with the Ca+2-sensitive fluorescence dye indo-1. FEBS Lett. 1989;256:55–61. doi: 10.1016/0014-5793(89)81717-x. [DOI] [PubMed] [Google Scholar]
- 31.Pozos TC, Sekler I, Cyert MS. The product of HUM1, a novel yeast gene, is required for vacuolar Ca2+/H+ exchange and is related to mammalian Na+/Ca2+ exchangers. Mol Cell Biol. 1996;16:3730–3741. doi: 10.1128/mcb.16.7.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller AJ, Vogg G, Sanders D. Cytosolic calcium homeostasis in fungi: roles of plasma membrane transport and intracellular sequestration of calcium. Proc Natl Acad Sci U S A. 1990;87:9348–9352. doi: 10.1073/pnas.87.23.9348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stroobant P, Scarborough GA. Active transport of calcium in Neurospora plasma membrane vesicles. Proc Natl Acad Sci U S A. 1979;76:3102–3106. doi: 10.1073/pnas.76.7.3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cornelius G, Nakashima H. Vacuoles play a decisive role in calcium homeostasis in Neurospora crassa. J Gen Microbiol. 1987;133:2341–2347. [Google Scholar]
- 35.Cramer CL, Davis RH. Polyphosphate-cation interaction in the amino acid-containing vacuole of Neurospora crassa. J Biol Chem. 1984;259:5152–5157. [PubMed] [Google Scholar]
- 36.Bowman BJ, Draskovic M, Freitag M, Bowman EJ. Structure and distribution of organelles and cellular location of calcium transporters in Neurospora crassa. Eukaryot Cell. 2009;8:1845–1855. doi: 10.1128/EC.00174-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bowman BJ, Abreu S, Margolles-Clark E, Draskovic M, Bowman EJ. Role of four calcium transport proteins, encoded by nca-1, nca-2, nca-3, and cax, in maintaining intracellular calcium levels in Neurospora crassa. Eukaryot Cell. 2011;10:654–661. doi: 10.1128/EC.00239-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Findon H, Calcagno-Pizarelli AM, Martínez JL, Spielvogel A, Markina-Iñarrairaegui A, et al. Analysis of a novel calcium auxotrophy in Aspergillus nidulans. Fungal Genet Biol. 2010;47:647–655. doi: 10.1016/j.fgb.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rispail N, Soanes DM, Ant C, Czajkowski R, Grünler A, et al. Comparative genomics of MAP kinase and calcium-calcineurin signalling components in plant and human pathogenic fungi. Fungal Genet Biol. 2009;46:287–98. doi: 10.1016/j.fgb.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 40.Yoshimoto H, Saltsman K, Gasch AP, Li HX, Ogawa N, et al. Genome-wide analysis of gene expression regulated by the calcineurin/Crz1p signaling pathway in Saccharomyces cerevisiae. J Biol Chem. 2002;277:31079–31088. doi: 10.1074/jbc.M202718200. [DOI] [PubMed] [Google Scholar]
- 41.Hagiwara D, Kondo A, Fujioka T, Abe K. Functional analysis of C2H2 zinc finger transcription factor CrzA involved in calcium signaling in Aspergillus nidulans. Curr Genet. 2008;54:325–338. doi: 10.1007/s00294-008-0220-z. [DOI] [PubMed] [Google Scholar]
- 42.Spielvogel A, Findon H, Arst HN, Araújo-Bazán L, Hernández-Ortíz P, et al. Two zinc finger transcription factors, CrzA and SltA, are involved in cation homoeostasis and detoxification in Aspergillus nidulans. Biochem J. 2008;414:419–29. doi: 10.1042/BJ20080344. [DOI] [PubMed] [Google Scholar]
- 43.Kafer E. Meiotic and mitotic recombination in Aspergilllus and its chromosomal aberrations. Adv Genet. 1977;19:33–131. doi: 10.1016/s0065-2660(08)60245-x. [DOI] [PubMed] [Google Scholar]
- 44.Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, et al. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci USA. 2006;103:10352–10357. doi: 10.1073/pnas.0601456103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schiestl RH, Gietz RD. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 46.Goldman GH, dos Reis Marques E, Duarte Ribeiro DC, de Souza Bernardes LA, Quiapin AC, et al. Expressed sequence tag analysis of the human pathogen Paracoccidioides brasiliensis yeast phase: identification of putative homologues of Candida albicans virulence and pathogenicity genes. Eukaryot Cell. 2003;2:34–48. doi: 10.1128/EC.2.1.34-48.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Herrera-Estrella A, Goldman GH, Van Montagu M. High-efficiency transformation system for the biocontrol agents, Trichoderma spp. Mol Microbiol. 1990;4:839–43. doi: 10.1111/j.1365-2958.1990.tb00654.x. [DOI] [PubMed] [Google Scholar]
- 48.Semighini CP, Marins M, Goldman MHS, Goldman GH. Quantitative analysis of the relative transcript levels of ABC transporter Atr genes in Aspergillus nidulans by Real-Time Reverse Transcripition-PCR assay. Appl Environ Microbiol. 2002;68:1351–1357. doi: 10.1128/AEM.68.3.1351-1357.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 50.Hartree EF. Determination of protein: a modification of the Lowry method that gives a linear photometric response. Anal Biochem. 1972;48:422–427. doi: 10.1016/0003-2697(72)90094-2. [DOI] [PubMed] [Google Scholar]
- 51.Mota Júnior AO, Malavazi I, Soriani FM, Heinekamp T, Jacobsen I, et al. Molecular characterization of the Aspergillus fumigatus NCS-1 homologue, NcsA. Mol Genet Genomics. 2008;280:483–95. doi: 10.1007/s00438-008-0381-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Clustal alignment of A. fumigatus PmcA, PmcB, and PmcC.
(DOCX)
(B) CrzA-binding regions of the pmcA-C genes (upstream the ATG start codon). Underline and in bold the putative CDRE-motif.
(DOCX)
Southern blot and PCR analyses for (A) ΔvcxA, (B) ΔpmcB, (C) ΔpmcA and (D) alcA::pmcC.
(PPT)
Histological analysis of alveolar lavages after infection with the A. fumigatus wild type, ΔcalA, and ΔcrzA strains. Germlings and host cells were detected by using Grocotts methenamine silver and haematoxylin and eosin staining, respectively. Bars, 100 µm.
(PPTX)
A. fumigatus ΔpmcB virulence studies in neutropenic mice. Comparative analysis of wild type and ΔpmcB strains in a neutropenic murine model of pulmonary aspergillosis. A group of 10 mice per strain was infected intranasally with 20 µl suspension of conidiospores at a dose of 2.0–5.0×104.
(PPTX)
(A) Growth phenotype of the ΔpmcA::pmcA+ strain grown on YAG+CaCl2 500 mM. (B) PCR of the pmcA open reading frame. (C) PCR of the pmcB open reading frame.
(PPT)
List of primers and probes used in this work.
(DOC)