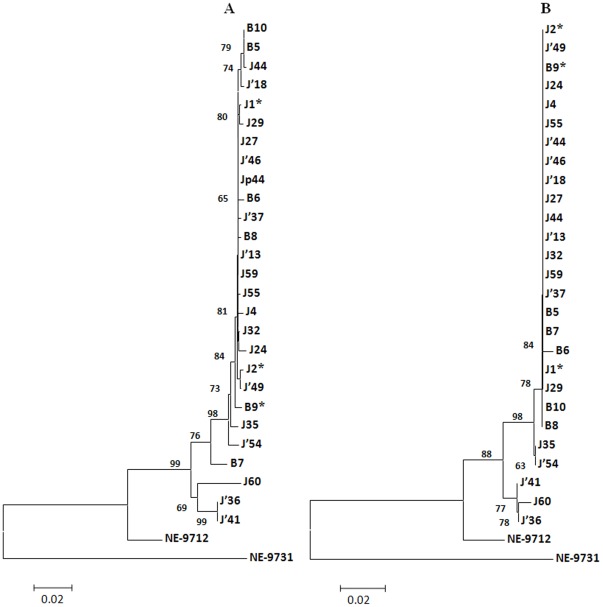
Figure 3. Phylogenetic trees for segment 10 and VP10 of LNV isolates from Xinjiang province.
A: Phylogenetic tree based on the nucleotide sequences. B: Phylogentic tree based on the amino acid sequences. Bootstrap values for 500 replications are indicated at each node. The trees were constructed using the Neighbor joining method and the Kimura 2 parameters method (nucleotides) and Poisson's correction method (amino acids). Similar trees were obtained with the P-distance algorithm. The bar represents the number of substitutions per site. Isolate designations are those reported in table 2. NE-9712 (LNV-NE9712) and NE-9731 (LNV-NE9731) are prototype strains of LNV serotypes 1 and 2, respectively. The letter B in isolates designation refers to Boshikeranmu while J refers to Jiashi county.