Fig. 2.
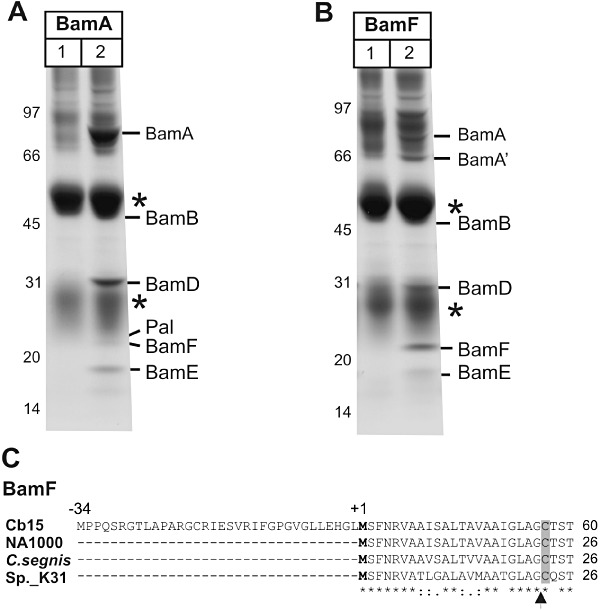
BamF is a subunit of the BAM complex in C. crescentus. A. Outer membrane vesicles were solubilized with 0.75% DDM and subject to immunoprecipitation using pre-immune serum (lane 1) or using BamA anti-serum (lane 2), and analysed by SDS-PAGE and Coomassie blue staining. Asterisks indicate the IgG heavy and light chains. Mass spectrometry was used to identify the indicated components of the BAM complex, and the novel protein BamF. B. Outer membrane vesicles were solubilized with 0.75% DDM and subject to immunoprecipitation using pre-immune serum (lane 1) or using BamF anti-serum (lane 2), and analysed as described above. BamA′ designates a 66 kDa protein fragment of BamA. C. The BamF sequences from C. crescentus strains CB15 and NA1000, Caulobacter segnis and Caulobacter sp. 31 are shown. The genome sequence from CB15 is unique in having an in-frame ATG codon (corresponding to Met ‘−34’) upstream of the predicted start site in all other Caulobacter genomes. The predicted processing sites for Signal peptidase II is indicated with an arrow-head. Asterisks indicate sequence identity across the alignment and the cysteine residue predicted to be lipid-modified is shaded grey.
