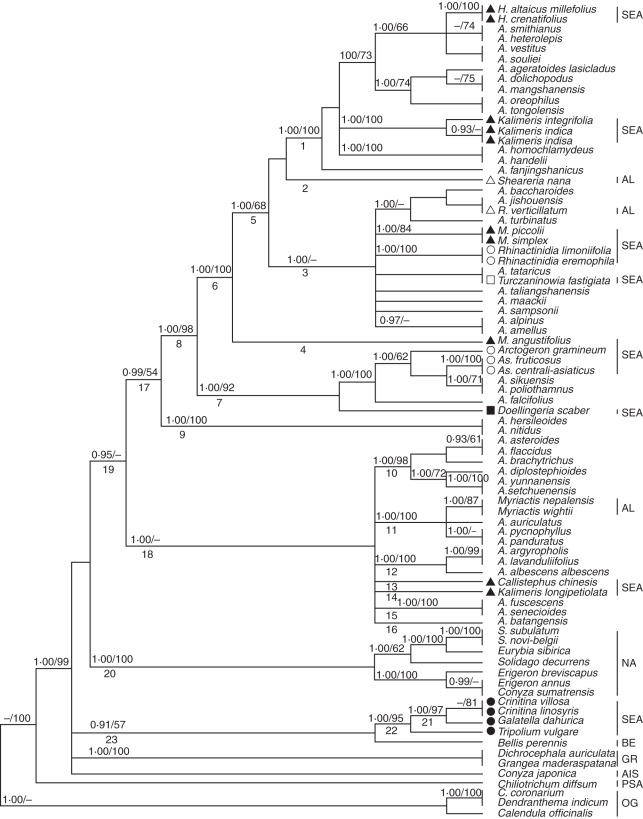
Fig. 2.
The 50 % majority rule consensus tree from the Bayesian analysis of the combined data set (nuclear ribosomal DNA internal and external transcribed spacer sequences and plastid genome DNA trnL-F sequences). Bayesian posterior probabilities (≥0·89) and bootstrap values (≥50 %) are indicated above the branches; ‘–’ indicates that Bayesian posterior probabilities are <0·89 or bootstrap percentages are <50 %. Some clades are indicated by numbers below the branch. Abbreviations: A., Aster; As., Asterothamnus; C., Chrysanthemum; H., Heteropappus; M., Miyamayomena; R., Rhynchospermum; S., Symphyotrichum. Abbreviations of the lineages are identical to those given in the Appendix and are shown on the right side of the taxa. Some species are labelled with the symbols shown in Fig. 1.