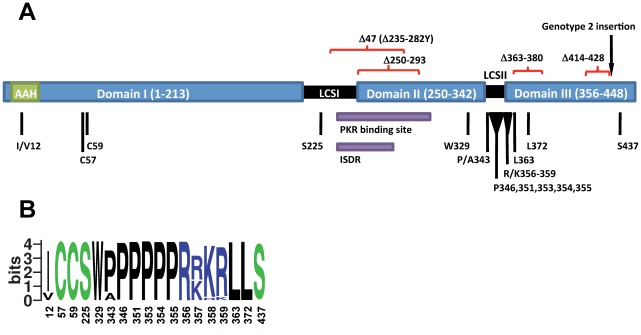
Figure 1. HCV NS5A residues and regions analyzed in reverse genetic studies.
Numbering is according to the H77 reference NS5A sequence (GenBank accession number AF009606). (A) Schematic drawing of the NS5A protein. The putative domains I–III (blue), are linked by LCSI and II (black). The amphipathic alpha-helix (AAH) in domain I is predicted to cover residues 5–25 (green) [18]. A putative IFN-sensitivity determining region (ISDR) covering residues 237–276 [25] and a PKR binding region covering residues 237–302 [24] (purple) extend from the LCSI into domain II. Positions mutated in reverse genetic studies are indicated in black and deletions are indicated in red. An NS5A amino acid alignment of isolates studied is shown in Figure S1. (B) Variation at positions mutated in reverse genetic studies among isolates in the most recent pre-made NS5A web-alignment consisting of 673 entries from the Los Alamos HCV sequence database visualized as logo plot [63]. The information content at every position is given in bits. Amino acids are colored by properties: polar (green), basic (blue), acidic (red) and hydrophobic (black). In addition to the residues shown, a few serines were found at residues 346 and 354, while a few asparagines were found at residue 437. Residues observed in less than two sequences were not included.