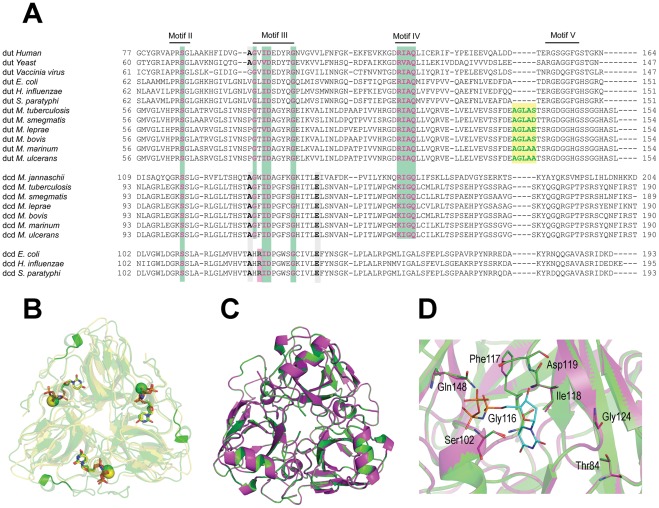
Figure 2. Sequence and structural comparison of selected members of the dUTPase superfamily.
(A) Conserved motifs are indicated above the sequences as lines. Representative organisms from widely different evolutionary branches are also included for comparison. Mycobacterial dCTP deaminases contain all those conserved residues that are indispensable for dUTPase reaction. Residues conserved between dUTPases and bifunctional dCTP deaminase/dUTPases are important for the dephosphorylation reaction and indicated with green boxes. Residues important for the deamination reaction and crucial for dCTP deaminase monofunctionality are depicted as gray and magenta boxes, respectively. Mycobacterial dUTPases contain an insert present solely in the mycobacterial dut, this insert is shown as a yellow box. The alignment was performed with ClustalW. (B) The mycobacterial insert induces a loop structure on the surface of the dUTPase monomer. The superimposed structure of hDUT (PDB ID: 3EHW, under publication in a separate paper) and mtDUT (PDB ID: 2PY4) are depicted as yellow and green cartoon representation, respectively. The mycobacterial insert can be seen as cartoon tube representation. In the active sites the bound ligands, dUPNPP and Mg2+ can be seen, whereby the Mg2+ is visualized as a yellow (hDUT) or green (mtDUT) sphere while dUPNPP is represented as sticks with atomic coloring (carbons in yellow and green). Structures were prepared using PyMol. (C) Superimposed overall structure of the M. tuberculosis bifunctional dCTP deaminase/dUTPase and the M. smegmatis dCTP deaminase enzymes in green and magenta cartoon representation, respectively. (D) Enlarged view from C showing the conserved residues and the non-hydrolysable substrate analog α-β-imido-dUTP (dUPNPP) as modeled to the active site. Residues and the dUPNPP molecule are in stick representation with atomic coloring (green, magenta and cyan carbons for M. tuberculosis, M. smegmatis enzymes and dUPNPP, respectively). Note the closely identical organization of both the overall structure and the active site in M. tuberculosis and M. smegmatis dUTPases.