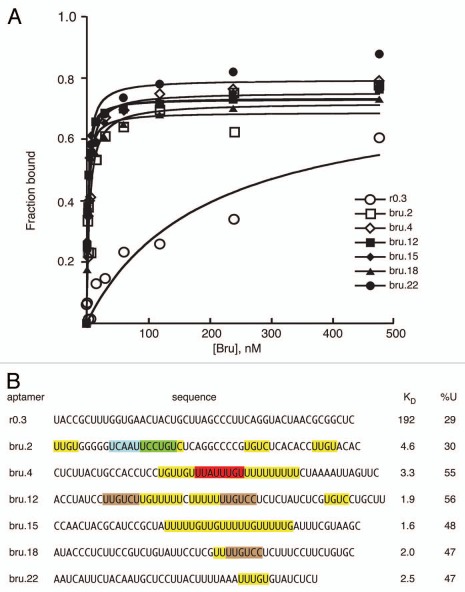
Figure 2.
Bru binding to Anti-Bru aptamers. (A) Results of filter binding assays with recombinant Bru (0.9–477 nM) and anti-Bru aptamers. (B) Aptamer sequences (excluding the sequences from the cloning vector that are common to all) are shown, with KD values obtained from the data of part A. Motifs identified from the in vitro selections are shaded. The predominant UUGUCY motif from the Bru selection is shaded brown, with only perfect copies indicated. All of the anti-Bru aptamers have at least one copy of a singly-mismatched version of the motif (these are not shaded), while the control RNA obtained from the pool prior to selection (r0.3) has none. Examples of the motifs from the RRM3+ selection that also appeared in the Bru selection are shaded in green and red. Instances of the 5 tetranucleotides that appear most frequently in the anti-Bru aptamers (UUGU, UUUU, UGUC, UUUG and UGUU; Sup. Table 4) are shaded in yellow (except where the sequences are within another shaded region). The proportion of U, which is highly enriched in the Bru selection, is given for each RNA.