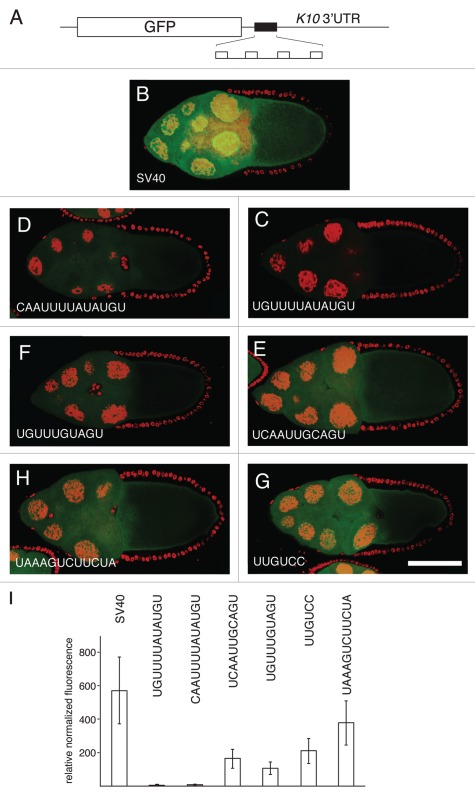
Figure 3.
Translational repression by candidate regulatory sites. (A) schematic diagram of reporter mRNAs with the variable region indicated by a filled box. For the SV40 reporter, the variable region is only SV40 sequences. For the remaining reporter mRNAs the variable region has four copies of a candidate Bru regulatory site embedded in SV40 sequences. (B–H) examples of GFP levels in stage 10A egg chambers expressing GFP reporter transgenes with the matα4-GAL-VP16 driver. All confocal images were taken on the same day at the same settings. The scale bar represents 75 µm. (B) control GFP transgene with no anti-Bru aptamer binding motifs. The remaining image parts are for GFP transgenes with the Bru binding motifs indicated in the figure and described below. (C) UGUUUUAUAUGU is from the osk AB region, and consists of a BRE-like motif adjacent to a short U/G rich motif (like those from the RRM1+2 selection). (D) CAAUUUUAUAUGU is from the cycA 3′UTR, and consists of a BRE-like sequence adjacent to a short C/A rich motif (like those from the RRM1+2 selection). (E) UCAAUUGCAGU is from the cycA 3′ UTR, and consists of a copy of the UGCAGU motif (from the RRM3+ selection) adjacent to a short C/A rich motif (like those from the RRM1+2 selection). (F) UGUUUGUAGU is from the grk 3′UTR, and consists of the UGCAGU motif (from the RRM3+ selection but with a single mismatch) adjacent to a short U/G rich motif (like those from the RRM1+2 selection). (G) UUGUCc is the type II Bru binding site, which appears three times in the AB and C regions of the osk 3′UTR. (H) UAAAGUCUUCUA is from the osk C region, and is a type III Bru binding site with a single mismatch relative to the longest aptamer motif from the RRM3+ selection. (I) Relative GFP levels in the nurse cell cytoplasm of stage 9/10 egg chambers for each of the reporter transgenes. GFP levels (obtained from 45 measurements for each transgene) were normalized to the RNA levels (from 3 measurements for each transgene). Transgene RNA levels were normalized relative to rp49 RNA levels. All of the reporter transgene mRNAs with candidate regulatory sites show reductions in GFP levels that are significantly lower than for the control (p < 0.0001 by the Tukey-Kramer method).