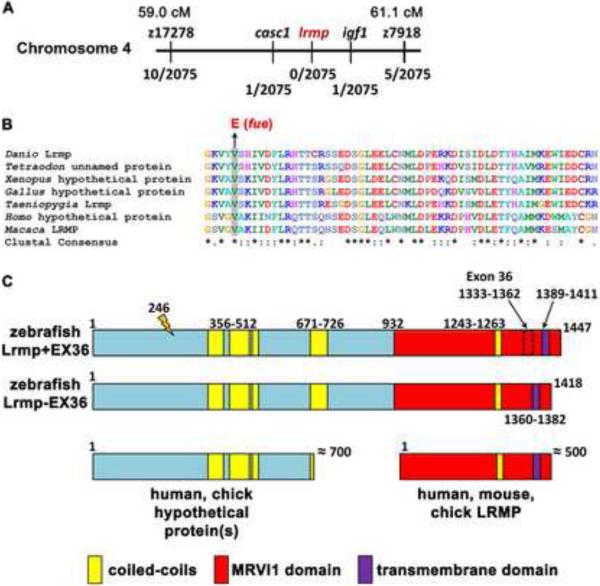
Figure 2. The fue molecular lesion results in an amino acid substitution at a conserved residue of lymphoid-restricted membrane protein.
(A) Positional cloning indicated linkage of fue to chromosome 4 between markers z17278 and z7918. On the physical map, the mutation lies between markers in cancer susceptibility candidate 1 (casc1) and insulin-like growth factor 1 (igf1). Fractions below markers indicate the number of recombinant meiotic events at each position out of 2075 examined meioses. (B) A point mutation was discovered in the fourth exon of lrmp, predicted to cause a valine-to-glutamate amino acid substitution at a conserved residue. (C) Alternative splicing involving exon 36 results in proteins of 1447 (Lrmp+EX36, NCBI accession #CAI20727) and 1418 (Lrmp−EX36) residues. The N-terminal region of zebrafish Lrmp is homologous to hypothetical proteins in humans, chick, and several other species. The C-terminal 500 residues in zebrafish Lrmp correspond to Lrmp proteins described in mouse and human cells, and predicted in chick. Predicted domains and fue mutation site (orange marker at amino acid 246) are indicated. See also Figure S2 and Table S1.