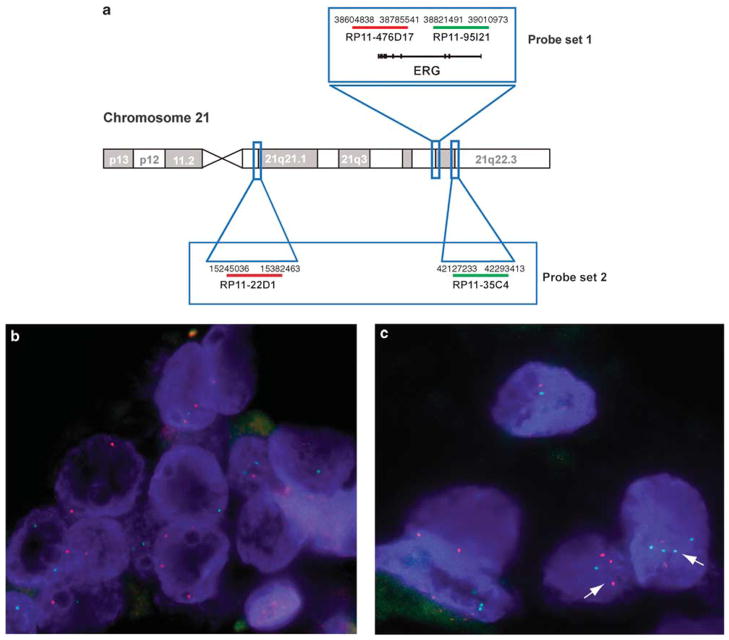
Figure 2.
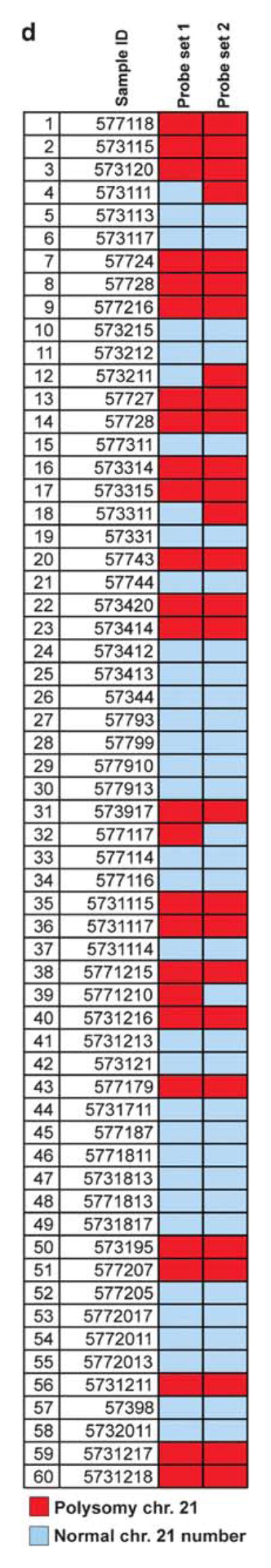
Evaluation of chromosome 21 numerical alterations. (a) Overview of BAC clones used in the study. Probe set 1 flanking the ERG genomic locus (RP11-476D17, RP11-95I21) is the set used to determine ERG gene rearrangement status in all cases in the current study. Probe set 2 was only used in a subset of cases to further assess chromosome 21 numerical alterations by targeting centromeric (RP11-22D1) and telomeric (RP11-35C4) regions on the long arm of chromosome 21. (b) Prostate adenocarcinoma showing no evidence of numerical chromosome 21 alteration using probe set 2. (c) Prostate adenocarcinoma showing chromosome 21 long-arm copy number gains as indicated by the presence of more than two red and/or more than two green signals per nucleus (white arrows). (d) Comparison of ERG gene copy number gain detected using probe set 1 and chromosome 21 numerical alteration assessed using probe set 2 in a subset of two tissue microarrays (60 cases) from our study. A high concordance rate (24/26; 93%) is found between the two sets of probes supporting that ERG gene copy number gain are a reflection of chromosome 21 copy number gains.