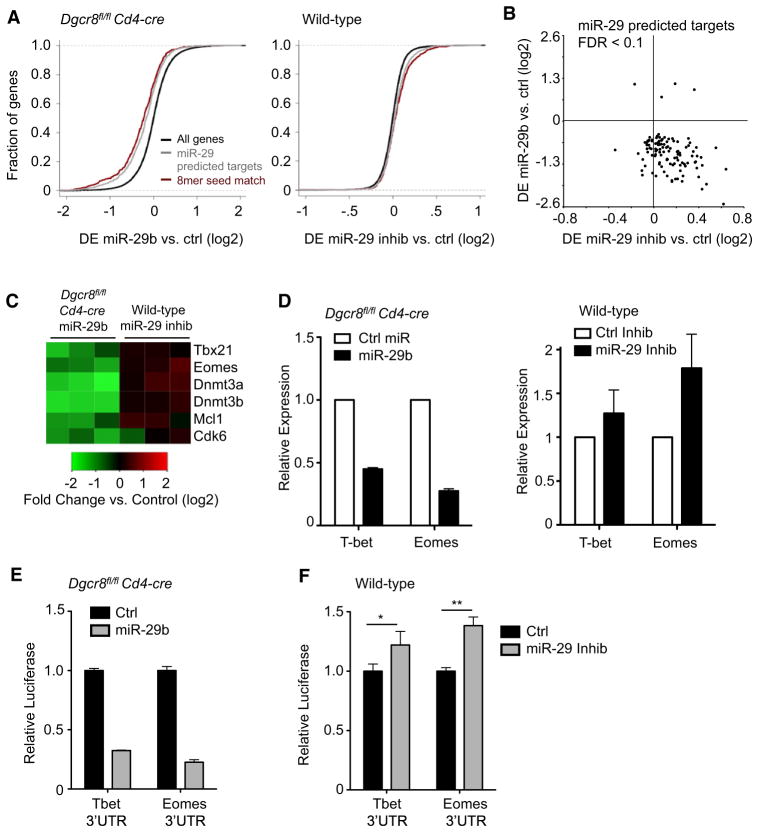
Figure 5. Genome-wide Analysis Identifies T-bet and Eomes as miR-29 Targets.
(A) Cumulative distribution of mRNA expression changes from microarray data subsequent to transfection with miR-29b in Dgcr8fl/fl Cd4-cre CD4+ T cells (left) or miR-29 inhibitor in C57BL/6 (wild-type) CD4+ T cells (right). Negative values indicate downregulation of mRNA after transfection. Black lines represent the entire microarray gene set, gray lines represent the subset of genes that are Targetscan computationally predicted miR-29 targets, and red lines represent the subset of predicted targets with 8-mer binding sites perfectly complementary to the entire miR-29 seed sequence.
(B) Scatter plot of the change in mRNA expression after miR-29 transfection in Dgcr8fl/fl Cd4-cre cells (y axis) and change in expression after miR-29 inhibitor transfection in wild-type cells (x axis). Each point represents a different gene array probe. Included are predicted miR-29 targets differentially expressed (FDR < 0.1) after miR-29 transfection.
(C) Heatmap representation of changes in gene expression after transfection for three independent biological samples of each genotype. Scale is log2 fold change in array hybridization intensity compared to control transfected cells.
(D) qPCR validation of array data for miR-29 candidate targets of interest. mRNA expression was normalized to Gapdh and is relative to expression in control transfected cells. Values are means ± SD from three biological replicates.
(E and F) Primary Dgcr8fl/fl Cd4-cre (E) or wild-type (F) CD4+ T cells were cotransfected with a dual luciferase reporter and miR-29b or control miRNA (E); miR-29 inhibitors or control inhibitor (F). Luciferase reporters contained the full-length mouse 3′UTR of Tbx21 or Eomes. Renilla luciferase activity was measured 24 hr after transfection and normalized to firefly luciferase activity. Values are relative to normalized luciferase in control transfected cells. Data are representative of three independent experiments and values are means ± SD from three independent transfections.
*p < 0.05, **p < 0.01; ANOVA Tukey’s post hoc test.