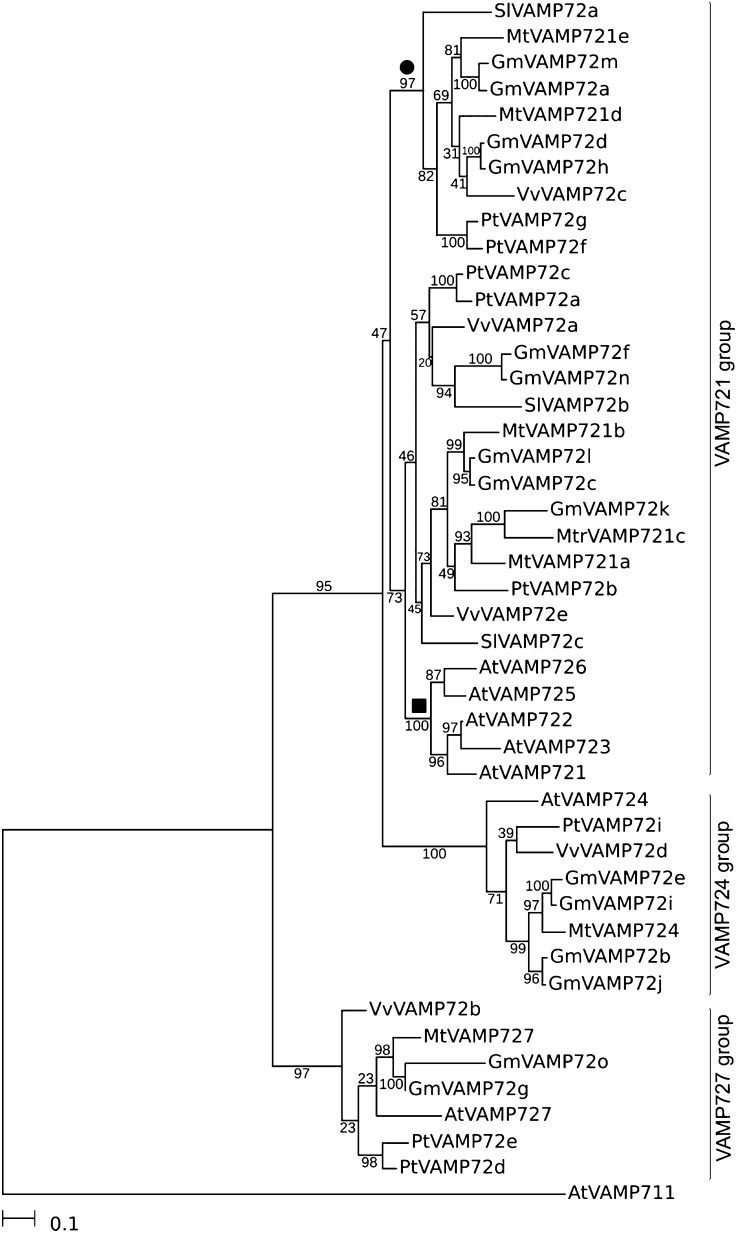
Fig. 3.
Phylogenetic analysis of MtVAMP72s. Unrooted phylogenetic neighbor-joining maximum-likelihood tree of M. truncatula (Mt) and Arabidopsis thaliana (At) Populus thrichocarpa (Pt), Glycine max (Gm), Solanum lycopersicum (Sl), and Vitis vinifera (Vv) VAMP72s. The sequences were identified using BLASTN searches in genome/EST databases and aligned at the protein level using MUSCLE v3.8.31. The phylogenetic tree was built using PHYML version 3.0 using protein-coding nucleotide sequences aligned using the protein alignment as a template. We used the general time-reversible model with γ-distributed rates of evolution with six categories. AtVAMP711 was used as outgroup, 100 bootstrap repetitions were performed to assess robustness of nodes. Note that MtVAMP721d and MtVAMP721e form a separate “symbiotic” subgroup (●, bootstrap 97%) inside VAMP721 group, whereas other MtVAMP721a, MtVAMP721b, and MtVAMP721c form other subgroup which also includes all Arabidopsis VAMP721 genes (■, bootstrap 73%). The latter are crucial in defense against fungal pathogens.