Abstract
Background
Ecological processes are increasingly being viewed as an important mode of diversification in the marine environment, where the high dispersal potential of pelagic organisms, and a lack of absolute barriers to gene flow may limit the occurrence of allopatric speciation through vicariance. Here we focus on the potential role of ecological partitioning in the diversification of a widely distributed group of marine protists, the planktonic foraminifera. Sampling was conducted in the tropical Arabian Sea, during the southwest (summer) monsoon, when pronounced environmental conditions result in a strong disparity in temperature, salinity and productivity between distinct northern and southern water masses.
Results
We uncovered extensive genetic diversity within the Arabian Sea planktonic foraminifera, identifying 13 morphospecies, represented by 20 distinct SSU rRNA genetic types. Several morphospecies/genetic types displayed non-random biogeographical distributions, partitioning between the northern and southern water masses, giving a strong indication of independent ecological adaptations.
Conclusions
We propose sea-surface primary productivity as the main factor driving the geographical segregation of Arabian Sea planktonic foraminifera, during the SW monsoon, with variations in symbiotic associations possibly playing a role in the specific ecological adaptations observed. Our findings suggest that ecological partitioning could be contributing to the high levels of 'cryptic' genetic diversity observed within the planktonic foraminifera, and support the view that ecological processes may play a key role in the diversification of marine pelagic organisms.
Background
The vast environment of the global ocean presents a challenge to the study of speciation. Marine planktonic microorganisms exist in huge populations, and carry a high passive dispersal potential [1]. With the presence of few physical barriers to gene flow in the open ocean, the occurrence of speciation through vicariant processes should be severely reduced, leading to large cosmopolitan, and genetically uniform populations [2,3]. Yet genetic data is increasingly highlighting the presence of "cryptic" diversity within many marine organisms [4-18], indicating that species diversity within the pelagic realm is significantly higher than suggested from many morphological taxonomies (reviewed in [2]). While vicariance clearly does play a role in the diversification of pelagic organisms [9,19], ecological speciation is increasingly being viewed as an important mode of diversification in the marine environment [2,20-22]. Here reproductive isolation can be achieved in the absence of intrinsic barriers to gene flow, by means of divergent selection for alternative environmental conditions or food resources [22-26]. Ecological partitioning has now been demonstrated to play a role in the speciation of a number of marine organisms [4,7-9,20,27-30], however, a great deal of further study will be necessary before the process can be fully understood.
Here we focus on the potential role of ecological partitioning in the diversification of the Planktonic Foraminifera, a highly diverse and widespread group of marine pelagic protists. The foraminifera are an important group, used frequently for paleoceanographic studies, and as a proxy for past climate change. Their utility is owed to an exceptional fossil record, spanning over 180 million years (Ma), and to the fact that individual "morphospecies" (identified by shell morphology) display characteristic environmental preferences, which are reflected in their spatial and temporal distribution in the oceans, and in the chemistry of their calcite shells. High levels of sequence variation have been found in the small subunit (SSU) ribosomal (r) RNA gene of the planktonic foraminiferal morphospecies, indicating the presence of numerous 'cryptic' genetic types [4-13,15], with mounting evidence indicating that these individual genetic types may display non-random geographical distributions, indicative of distinct ecological adaptations (ecotypes) [4-13,15].
Genetic surveys of the planktonic foraminifera have been undertaken over a wide range of oceanic water masses [4-6,8,10,11,13,22,31-36], though these ranged largely towards the mid to higher latitudes, with the species-rich tropics [37] remaining relatively under-sampled by comparison. Studies of high latitude planktonic foraminifera indicate that both vicariant [6-9,38,39] and ecological [4,7-9] processes may play a role in their diversification. Vicariance is implied by the presence of isolated or endemic genetic types within some morphospecies, likely resulting from the presence of physical barriers, such as the shallow Bering and Chukchi seas [8], or from oceanographic barriers such as the tropics and subtropics [4,8]. However, Darling and Wade [9] concluded that ecological constraints appeared to be major drivers of divergence in planktonic foraminifers in the high latitudes and anticipated that ecological factors would prove to be of prime importance in diversification in the mid to lower latitudes, where vertical niche partitioning is thought to be the principle factor controlling the distribution of foraminiferal morphospecies diversity [37].
For this study, the Arabian Sea was chosen as a tropical region of high priority. This unique marine environment is one of the richest marine biological areas in the world, and harbours a broad range of planktonic foraminiferal morphospecies [40]. It is subject to greater seasonal variability than any other ocean basin on the globe [41,42], with seasonally reversing monsoon winds inverting its circulation completely on a biannual basis [43,44]. In the winter months (November - February) prevailing winds progress in a northeasterly direction (the northeast monsoon), while in the summer months (June - September) they progress in a southwesterly direction (the southwest monsoon). During the summer monsoon, the formation of a major low-level air current, the Findlater jet [45], promotes upwelling in the coastal regions of Somalia, Yemen, and Oman [46], bringing nutrients into the euphotic zone. An enormous increase in primary productivity in the region results [47,48], transforming the normally nutrient poor (oligotrophic) waters of the northern Arabian Sea into one of the most productive (eutrophic) marine environments on Earth. At the same time, current circulations prevent the effect of this influx extending to the southern reaches of the Arabian Sea, which remain low in nutrients.
The Arabian Sea has been the focus of a number of studies linking physical oceanographic conditions to the distribution of planktonic foraminiferal morphospecies [49-52], however, this is the first time that the genetic diversity of the foraminifera within this region has been examined. This study investigates the biogeographical distributions of planktonic foraminiferal SSU rRNA genetic types in the Arabian Sea mixed layer during the SW (summer) monsoon, when pronounced environmental conditions lead to a distinct disparity in temperature, salinity and productivity between adjacent northern and southern water masses. Our results reveal non-random biogeographical distributions in several planktonic foraminiferal morphospecies/genetic types within the Arabian Sea during the SW monsoon, providing clear evidence of ecological partitioning.
Methods
Cruise track and oceanographic setting
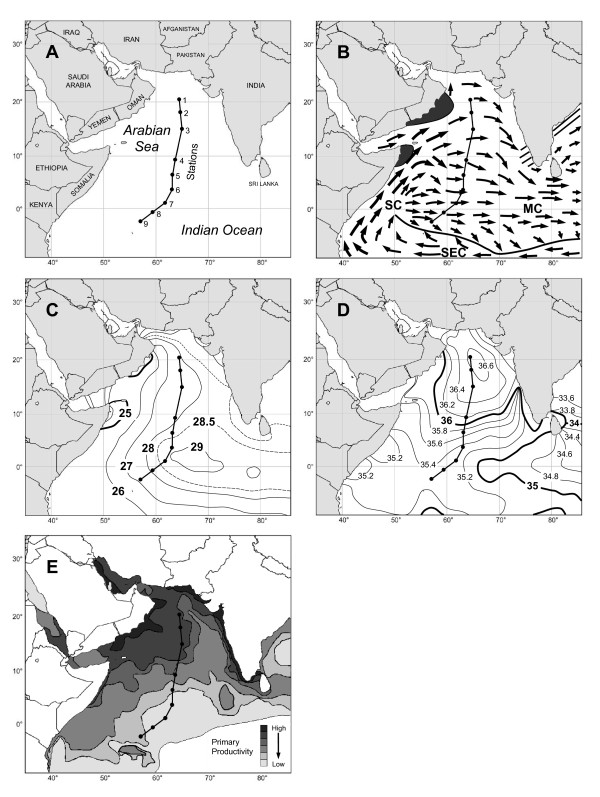
Specimens of planktonic foraminifera were collected at nine stations along a north/south cruise transect in the central Arabian Sea (20°22.81 N/64°29.36E-02°36.03 S/56°54.75E) during the summer monsoon of late June/July 2003 (Figure 1A; cruise Charles Darwin CD148, NERC). The oceanography of the Arabian Sea during the SW monsoon is shown in Figure 1B-E. Cyclonic surface circulation during the SW monsoon drives an eastward flowing monsoon current (MC) north of 10°S across the equatorial region (Figure 1B). A temperature gradient forms from west to east (Figure 1C) and there is a clear north/south differentiation in salinity (Figure 1D). Levels of primary productivity are elevated in the north of the Arabian Sea, but remain low in the oligotrophic south, with a water mass interface around stations 3-4 of the cruise transect (Figure 1E). Conductivity, temperature, depth (CTD) profiles from station 3 (15°01.11 N/65°00.02E) indicate that the mixed layer was 75 m deep at this position with a temperature of 28.5°C and a salinity of 36.7 psu, consistent with the maps in Figure 1C and 1D. The thermocline dipped steeply between 75 and 150 m (19°C) and then reduced its steepness coincident with a salinity minimum of 35.7 psu. Projections of mixed layer depth in July from Prasanna Kumar & Narvekar [53] indicate a mixed layer depth of ~50 m north of station 3, shoaling to a 40 m mixed layer depth south of station 5.
Figure 1.
Maps of the Arabian Sea showing the cruise transect and environmental conditions during the SW monsoon. (A) CD148 cruise transect and stations, (B) Surface currents during August at the SW monsoon peak. Regions of intense seasonal upwelling (dark grey), weak sporadic upwelling (hatched) SC = Somali Current, MC = Monsoon Current, SEC = Southern Equatorial Current (modified from [50]), (C) Average sea-surface temperature for the SW Monsoon in July 2005 (adapted from [54]), (D) Average sea-surface salinity for the SW Monsoon in July 2005 (adapted from [55]), (E) Average primary productivity during the SW monsoon in July - September 1979 (adapted from Coastal Zone Colour Scanner composite images of the region, NASA Earth-Sun System Division, Earth Sciences (GES) Data and Information Services Center (DISC) Distributed Active Archive Center (DAAC)).
Planktonic foraminiferal sampling
Samples were collected by pumping (5 m depth) from the ships' non-toxic water supply through a plankton screen (83 μm mesh) or by vertical net tow (0-100 and 0-200 m depth, 83 μm mesh) in waters with an average depth of 3,500 m. For genetic analysis, a representative sample of specimens was collected at each station. Individual specimens were identified using a stereomicroscope, and morphotype and cytoplasmic colouration were recorded by digital video imaging. Only adult specimens containing cytoplasm were selected for genetic analysis. These were crushed in a lysis buffer [56] and incubated for 1 hour at 60°C, before being transported to the lab where they were stored at -80°C. For assemblage assessment, bulk samples were taken at each station with the specimens either dried on slides directly or collected as bulk samples in ethanol. The preserved assemblages were then individually picked and placed onto micropalaeontological slides. The high incidence of small juveniles compared to the low incidence of mature specimens made identification too uncertain to carry out relative abundance counts along the transect, however, visual assessment of the bulk assemblages was undertaken.
PCR amplification and sequencing
The PCR amplification of an approximately 1,000 bp region of the terminal 3' end of the foraminiferal SSU rRNA gene was carried out using a nested PCR approach. 3 μl of template were used in the first round of PCR, using primer C5 coupled with either primer 138 or NS8 (Table 1). 1 μl of product from the first round was used as the template in the second round, initially using primers 2082F and 2514R (Table 1) for the identification of genetic types. For sequences found to be novel to the Arabian Sea, an ~1,000 bp fragment was amplified using primers 2082F and 3014R (Table 1) for use in phylogenetic tree reconstruction. PCR amplification was performed using 1 unit of Taq polymerase (Qiagen) or VentR polymerase (New England BioLabs) dependent upon success, with 200 μM each primer, 0.2 μM dNTPs, and 1.5 mM magnesium chloride in a 50 μl final volume. Thermal cycling (with a Perkin Elmer cycler) was performed with cycling parameters of 96°C for 2 mins, followed by 35 cycles of 96°C for 30 sec, 55°C for 30 sec and 72°C for 2 mins. Amplification products were purified from an agarose gel using a QIAprep spin miniprep (Qiagen). For taxa where direct sequencing was impossible due to the presence of multiple templates, cloning of the 1,000 bp fragment was carried out prior to sequencing using the TOPO TA® method (Invitrogen). Both sense and antisense strands were sequenced directly on an Applied Biosystems 377 DNA sequencer using BigDye terminator cycle sequencing.
Table 1.
SSU rRNA primer sequences
Sequence analysis
Sequences were assembled using Gap4 in the Staden package [59] and then aligned manually within version 2.2 of the Genetic Data Environment (GDE) package [60]. 90 foraminiferal taxa were selected for use in the main phylogenetic analysis, including all species/genetic types obtained from the Arabian Sea, together with examples of every species and genetic type of planktonic foraminifera currently available in GenBank, plus a representative group of benthic foraminifera (1 per family in GenBank, see Additional file 1). Great care was taken during the process of sequence alignment and in the selection of sites for use in subsequent analyses. The unusually high levels of sequence divergence observed among foraminiferal taxa in their rRNA genes makes the selection of unambiguously aligned sites for use in phylogenetic analysis particularly challenging. To ensure the accuracy of our phylogenetic analyses we adopted a conservative approach, utilising only those sites for which positional homology across all taxa was certain. In total, 407 base pairs (bp) could be unambiguously aligned across all foraminiferal taxa. To improve resolution, additional phylogenies were constructed for four of the most common Arabian Sea morphospecies, thus allowing a greater number of unambiguously aligned sites to be recruited into the analyses (Globigerinella siphonifera/Globigerinella calida (668 bp), Globigerinoides ruber/Globigerinoides conglobatus (589 bp), Globigerina bulloides (669 bp), Turborotalita quinqueloba (748 bp)). DNA sequence alignments are shown in Additional file 2 and Additional file 3.
Phylogenetic trees were constructed using Bayesian inference (BI) [61,62], maximum likelihood (ML) [63]), neighbour-joining (NJ) [64] and maximum parsimony (MP) [65]. BI was performed using the MrBayes (version 3.1.2) package [61] with multiple hits accounted for using a GTR + Γ model [66,67] and with the tree space explored using four chains of a Markov Chain Monte Carlo algorithm for 5 million generations (1 million for subset analyses), sampling every 100 generations. The run was terminated only after the Bayesian MCMC searches had reached a stationary phase (plateau), indicating convergence of the chain onto the target distribution, and a consensus tree built using the last 1000 trees (burn-in = 49001 samples for main tree, 9001 samples for subset analyses). ML analysis was undertaken within the Phyml package [68] using a GTR + Γ model [66,67], with parameters estimated within Phyml. NJ and MP analyses were performed using PAUP* (version 4.0d65; [69]). For NJ, distances were corrected using a GTR + Γ model [66,67] with the rate matrix, base frequencies, and shape parameter (α) of the gamma distribution (based on 16 rate categories) estimated using likelihood by iteration from an initial NJ tree. Bootstrap resampling [70] was undertaken using ML, NJ and MP with 1000 bootstrap replicates in order to assign support to particular branches within the tree. Bayesian posterior probabilities were obtained within MrBayes from the last 1000 trees generated.
The planktonic foraminiferal SSU rDNA sequences presented in this study are deposited in GenBank, accession numbers JQ799892 to JQ799900.
Results
363 specimens of planktonic foraminifera were collected from 8 stations along a cruise transect in the Arabian Sea during the summer monsoon of 2003 (Figure 1A). Small subunit rRNA gene sequences were successfully amplified for 213 individual specimens. Examination of the SSU rDNA sequences revealed high levels of genetic diversity within the Arabian Sea mixed layer planktonic foraminiferal population, with 20 different genetic types being recognised from 13 different morphospecies
Phylogenetic placement of the Arabian Sea foraminiferal genetic types
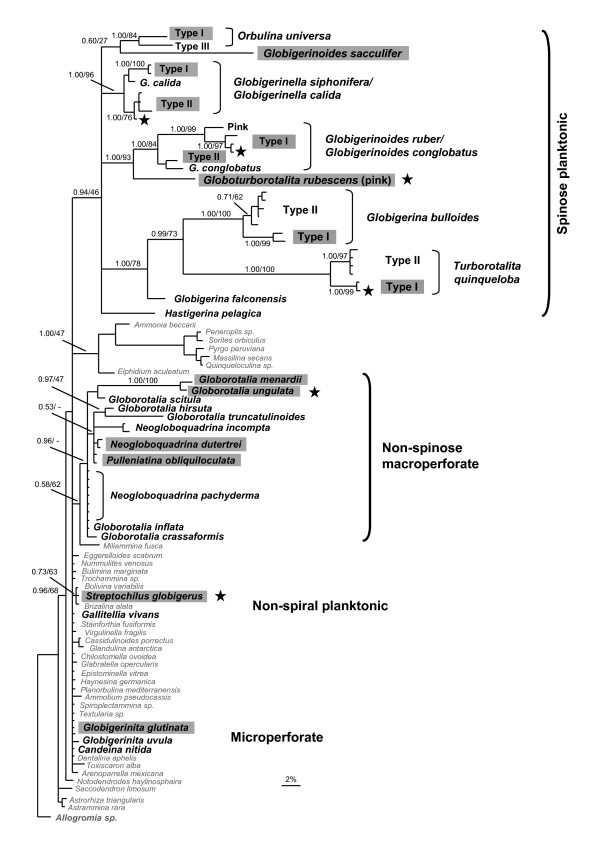
A comprehensive foraminiferal phylogeny, based on 407 bp of the SSU rRNA gene (Figure 2) highlights the placement of the Arabian Sea taxa. All methods of phylogeny reconstruction utilised were largely consistent in their inferred trees, and the phylogeny is in general agreement with previous studies [4,5,7,10,22,35]. The planktonic foraminifera appear polyphyletic, falling in at least 4 separate areas of the tree (Figure 2), consistent with the morphological groupings of the spinose (Globigerinidae and Hastigerinidae), non-spinose macroperforate (Globorotaliidae & Pulleniatinidae), non-spinose microperforate (Candeinidae), and the non-spiral planktonic foraminifera (see [71]).
Figure 2.
Bayesian inference SSU rDNA phylogenetic tree showing the position of the Arabian Sea morphospecies and genotypes within the foraminifera. The phylogeny is based on 407 unambiguously aligned nucleotide sites and is rooted on the benthic foraminifer Allogromia sp. Bayesian posterior probabilities (from the last 1000 trees, obtained within MrBayes) and ML bootstraps (expressed as a percentage, 1000 replicates) are shown on the tree (BI posterior probabilities/ML bootstraps). The scale bar corresponds to a genetic distance of 2%. Benthic foraminiferal taxa are shown in grey text, and planktonic foraminifera are shown in black. Morphospecies and genotypes found in the Arabian Sea are shown on a grey background. A star indicates a novel sequence obtained from the Arabian Sea cruise (CD148). The sequence for S. globigerus is also presented in [72].
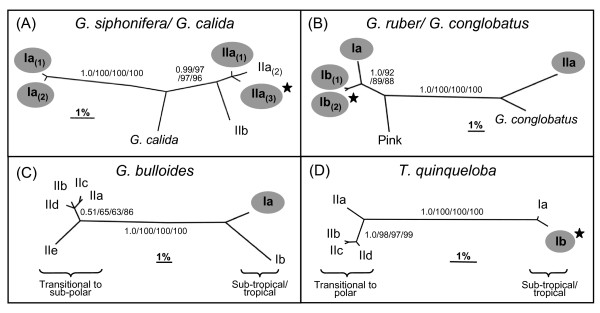
The spinose planktonic foraminifera were represented by seven morphospecies within the Arabian Sea mixed layer (Orbulina universa, Globigerinoides sacculifer, Globigerinella siphonifera, Globigerinoides ruber (white), Globoturborotalita rubescens (pink), Globigerina bulloides, and Turborotalita quinqueloba). Only a single O. universa Type I individual was genotyped, falling together with G. sacculifer (Figure 2). Four genetic types of G. siphonifera were identified (Types Ia(1), Ia(2), IIa(1), and the novel IIa(3)) (668 bp SSU rDNA phylogeny; Figure 3A). The subtle G. siphonifera Type IIa sub-types, shown previously as the IIa complex by Darling and Wade [9], are named here as subtypes IIa(1) (Genbank:U80788), IIa(2) (Genbank:AF102227, Genbank:AJ3905674, Genbank:Z83960), and IIa(3) (this study). Globoturborotalita rubescens (pink) is included in a foraminiferal phylogeny for the first time and falls in a well-supported clade as the sister taxon to G. ruber/G. conglobatus (Bayesian posterior probability (pp) = 1.00, 93% ML bootstrap support) (Figure 2). Globigerinoides ruber (white) was represented by four genetic types (Types Ia, Ib(1), the novel Ib(2), and IIa) (589 bp SSU rDNA phylogeny; Figure 3B). A subtly different variant of G. ruber Type Ib was discovered in the Arabian Sea, splitting Ib into subtypes Ib(1) and the new Ib(2). Globigerina bulloides was represented by Type Ia, which falls as a sister to Type Ib (669 bp SSU rDNA phylogeny; Figure 3C). A new variant of T. quinqueloba Type I was discovered, though only a single individual was successfully sequenced. It is named here as Type Ib and falls together with Type Ia (748 bp SSU rDNA phylogeny; Figure 3D).
Figure 3.
SSU rDNA phylogenetic trees of (A) Globigerinella siphonifera/Globigerinella calida (668 unambiguously aligned nucleotide sites), (B) Globigerinoides ruber/Globigerinoides conglobatus (589 bp), (C) Globigerina bulloides (669 bp), (D) Turborotalita quinqueloba (748 bp). The phylogenies were constructed using Bayesian Inference and are unrooted. Bayesian posterior probabilities and ML, NJ, and MP bootstraps (expressed as a percentage) are shown on the trees (BI/ML/NJ/MP). The scale bar corresponds to a genetic distance of 1%. Morphospecies and genotypes found in the Arabian Sea are shown on a grey background. A star indicates a novel sequence obtained from the Arabian Sea cruise (CD148).
Four non-spinose macroperforate morphospecies were present in the Arabian Sea (Globorotalia menardii, Globorotalia ungulata, Neogloboquadrina dutertrei, and Pulleniatina obliquiloculata) (Figure 2). Globorotalia menardii and the newly sequenced G. ungulata fell together (pp = 1.00 BI, 100% ML, Figure 2) with the other macroperforates, though the placement of G. menardii was inconsistent across tree reconstruction methods, possibly a result of its unusually high rate of evolution. Very minor sequence variation was detected in G. menardii, though insufficient to warrant sub-type status. The three specimens of G. ungulata exhibited the discriminating morphological features of this morphospecies (e.g. a keel structure on the umbilical shoulder of the test [71]), though some workers believe G. ungulata to be an immature form of G. tumida. For N. dutertrei, minor sequence variation was detected in the most variable regions of the SSU gene, as noted in other neogloboquadrinid morphospecies [9], however, extensive cloning would be required to determine whether individual genetic types are present. All Pulleniatina obliquiloculata sequences obtained were identical to each other, however differed subtly from those currently in GenBank. Further investigation will be necessary to determine if they represent a genetic sub-type of the species.
Of the three microperforate planktonic morphospecies sequenced to date; Globigerinita uvula [35], Globigerinita glutinata [22] and Candeina nitida [36], only G. glutinata was found in the Arabian Sea mixed layer. Examining all G. glutinata sequences available to date, three subtly different genetic types can be identified, named here as Type 1a(1) (Genbank:AF250105; and Arabian Sea) 1a(2) (Genbank:Z83974), and 1a(3) (Genbank:AY453136). Recent cloning of G. glutinata from North-West Pacific assemblages [36] indicates that these are most likely to be genuine subtype differences.
The non-spiral morphospecies, Streptochilus globigerus, one of two extant biserial planktonic foraminifera, was abundant in the Arabian Sea mixed layer and falls together with infaunal benthic biserial species Bolivina variabilis in the phylogeny (Figure 2; [72]). The morphospecies exhibits minor intra-specific variation in the SSU sequences, as in other benthic foraminiferal species [9].
Biogeographical distributions of the Arabian Sea foraminiferal genetic types
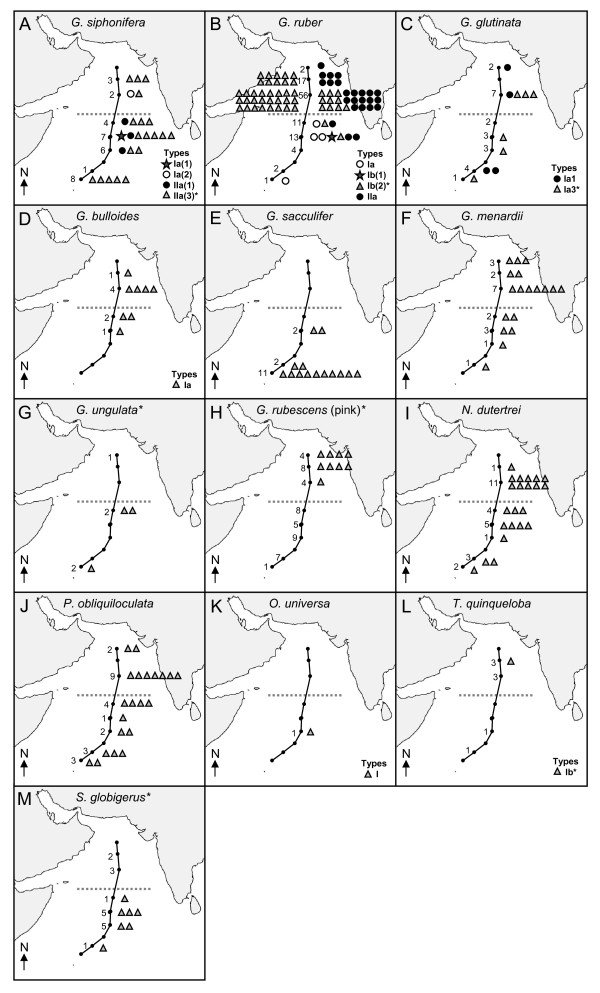
Analysis of the morphospecies genetic type distribution data (Figure 4) combined with a visual assessment of the bulk assemblage data showed some distinct ecological segregation related to the physical oceanography of the Arabian Sea.
Figure 4.
Arabian Sea maps showing the spatial distribution of genetic types of the morphospecies identified along the cruise transect. Their numbers do not necessarily reflect the absolute frequency of morphospecies or genotypes in the water column. The dotted line denotes approx. water mass boundary (higher productivity, high salinity in the north/oligotrophic, low salinity in the south) as determined from Figure 1D & 1E. The number of specimens collected at each station for genetic analysis is indicated by a number. A key to genotypes is provided on each map. * indicates novel sequences obtained from the Arabian Sea cruise (CD148).
Globigerinella siphonifera
Globigerinella siphonifera was distributed throughout the transect and was represented by four genetic types (Figure 4A). The newly recognised Type IIa(3) (n = 19) was distributed throughout the cruise transect, thriving equally in both the northern and southern water masses. The other genetic types appeared more rare. Type Ia(2) (n = 1) was found only in the northern water mass and Types Ia(1) (n = 1) and IIa(1) (n = 3) were found in low numbers in the southern water mass.
Globigerinoides ruber
Assessment of the bulk assemblage samples revealed that G. ruber was the dominant morphospecies in the Arabian Sea during the SW monsoon. It was found in high numbers in the more eutrophic, high salinity water of the north and occurred in significantly lower numbers in the more oligotrophic lower salinity water mass to the south. There are four genetic types of G. ruber in the Arabian Sea assemblage (Figure 4B), which have distinctive biogeographies. Only Type IIa (n = 24) and Type Ib(2) (n = 46) were found in the more eutrophic, higher salinity water mass of the northern Arabian Sea. The other G. ruber genetic types, Ia (n = 4) and Ib(1) (n = 1), were not found in the northern water mass following extensive genotyping of the water column. These genetic types were found in low numbers within the southern water mass, with only a single specimen of G. ruber Type Ia identified at station 9.
Globigerinita glutinata
Globigerinita glutinata was found throughout the transect. Two potentially distinct subtypes of Type Ia were identified in the central Arabian Sea mixed layer; Types 1a(1) (n = 4) and 1a(3) (n = 6), each distributed along the length of the cruise transect (Figure 4C).
Globigerina bulloides
Globigerina bulloides was present in very low numbers in the bulk samples and was distributed mainly in the more eutrophic, high salinity water mass of the northern region. Only genetic type Ia (n = 8) was found, confined to the northern water mass (Figure 4D).
Globigerinoides sacculifer
Globigerinoides sacculifer was found only in the southern waters, south of station 4. Only a single genetic type was found (n = 14), which was identical to all other G. sacculifer sequenced to date (Figure 4E).
Globorotalia menardii
The bulk assemblage data clearly showed that G. menardii was present across the whole transect with numbers increasing significantly towards the South, though this pattern was not reflected in the number of specimens collected for genotyping (Figure 4F). Despite the distribution difference between the water masses, only a single genetic type (n = 18) was found in the mixed layer along the cruise transect.
Globorotalia ungulata
Assessment of the bulk assemblage showed that Globorotalia ungulata was more common in the southern part of the cruise transect. Only three specimens were sequenced and a single genetic type found (n = 3; Figure 4G).
Globoturborotalita rubescens (pink)
Globoturborotalita rubescens (pink) was present throughout the transect, though only nine specimens were successfully amplified (Figure 4H). This newly sequenced morphospecies showed no sequence variation in the specimens collected between stations 1-3.
Neogloboquadrina dutertrei
Neogloboquadrina dutertrei was distributed along the length of the cruise transect, and is most likely represented by a single genetic type in the Arabian Sea (n = 22; Figure 4I). However, as in most Neogloboquadrina, N. dutertrei specimens exhibit intra-individual variation in their SSU gene repeats and the presence of more than one genetic type cannot be ruled out without extensive cloning.
Pulleniatina obliquiloculata
Pulleniatina obliquiloculata was distributed along the length of the cruise transect. Only a single genetic type was found (n = 21; Figure 4J).
Orbulina universa
Orbulina universa was very rare in the water column. Only a single specimen of Type I was identified in the southern water mass at station 6 (Figure 4K).
Turborotalita quinqueloba
It is difficult to differentiate T. quinqueloba from tiny juveniles of other morphospecies, but mature specimens were rare. Only a single specimen of Type Ia was amplified at station 2 (Figure 4L).
Streptochilus globigerus
Assessment of the bulk assemblage showed that the biserial morphospecies, S. globigerus, occurred in substantial numbers along the length of the cruise transect. Only a single genetic type was identified (n = 7; Figure 4M).
Discussion
Sampling of the tropical Arabian Sea during the SW monsoon uncovered a wealth of planktonic foraminiferal diversity. The 13 morphospecies found displayed high levels of SSU rRNA genetic diversity, with a total of 20 independent genetic types being recorded between them.
Three morphospecies: Globoturborotalita rubescens (pink), Globorotalia ungulata and Streptochilus globigerus were sequenced for the first time from Arabian Sea cruise CD148. Globoturborotalita rubescens (pink) falls at the base of a well-supported cluster with G. ruber and G. conglobatus (Figure 2). Fossil record studies show that it first appeared in the Middle Pliocene, around 3.6 million years ago [73] and may have evolved from Globigerina woodi [74] via the morphospecies Globigerina decoraperta [73]. Globorotalia ungulata falls together with the morphologically similar species, G. menardii, at the end of a relatively long branch in the main phylogeny (Figure 2). It appeared in the Late Pliocene around 2.5 million years ago and is thought to have evolved from Globorotalia tumida [73], however other extant globorotaliid morphospecies will need to be sequenced before their exact ancestry can be determined. Streptochilus globigerus fell among the benthic foraminifera in the main phylogeny (Figure 2), exhibiting extremely high sequence identity to the benthic species Bolivina variabilis, sufficient to suggest that S. globigerus and B. variabilis are one and the same morphospecies (discussed in more detail in [72]). In addition, four new foraminiferal genetic types (G. ruber Type Ib(2), G. siphonifera Type IIa(3), T. quinqueloba Type Ib and G. glutinata Type 1a(1)) were identified from this Arabian Sea cruise.
Evidence for ecological partitioning among the Arabian Sea morphospecies/genetic types
The varied hydrographic conditions and extreme seasonal variation of the Arabian Sea provide a unique environment within which to study the ecological adaptations of planktonic foraminiferal morphospecies and their individual genetic types. The cruise transect was conducted during the SW monsoon, when environmental conditions were most pronounced and a strong disparity existed between a high salinity, more eutrophic water mass in the north (Figure 1A-E; stations 1-3), and a low salinity, oligotrophic water mass in the south (Figure 1A-E; stations 4-9). Analysis of the spatial distributions of planktonic foraminiferal morphospecies and genetic types within the Arabian Sea mixed layer at this time revealed a number of non-random patterns of geographical distribution, suggestive of distinct ecological adaptations.
The spinose morphospecies, Globigerinella siphonifera (Figure 4A) and Globigerinoides ruber (Figure 4B), appear to offer particularly excellent examples of divergent biogeographies in their genetic types, each being represented by four individual genetic types, exhibiting apparently ecologically distinct distribution patterns.
Globigerinella siphonifera is represented by two highly divergent SSU rRNA genetic lineages, Type I and Type II, which from a wealth of biological evidence may be considered as two distinct species [4,15,75]. The newly recognised Type IIa(3) was the dominant genetic type of G. siphonifera in the Arabian Sea, and was distributed throughout both water masses in large numbers (n = 19) (Figure 4A), suggesting a broad tolerance for the varying hydrographic conditions. This new genetic type has yet to be found elsewhere, but may eventually be discovered in other parts of the Indo-Pacific, a region that has not been sampled extensively. Type IIa(1), conversely, was found only in small numbers in the southern water mass (n = 3), suggesting that despite the low level of genetic distinction (Figure 3A), Type IIa(1) may have more specialised ecological requirements than Type IIa(3). The closely related genetic types Ia(1) and Ia(2), represented by only single individuals, also displayed divergent ecologies, the former appearing in the oligotrophic southern water mass and the latter in the more eutrophic north. It is interesting to note that the main ecological divide between genetic types does not reflect their phylogenetic separation into the Type I and Type II lineages. Ecological partitioning instead appears to play a greater role in the divergence of closely related genetic types.
The SSU rRNA phylogeny of G. ruber is again characterised by a deep divergence between two extant lineages (lineage 1: G. ruber (white) types Ia, Ib(1), Ib(2), and G. ruber (pink), lineage 2: G. ruber (white) Type IIa and G. conglobatus) (Figure 3B), indicative of a species level distinction [5,9,31]. The biogeographical distribution of G. ruber genetic types in the Arabian Sea was unmistakably correlated to the hydrographic provinces during the SW monsoon (Figure 4B). Globigerinoides ruber dominated the more eutrophic/higher salinity water mass of the northern Arabian Sea, though genotyping revealed the presence of only two of the G. ruber genetic types here (Ib(2) and IIa), both occurring in equally high numbers. The other two genetic types (Ia and Ib(1)) were absent from the more eutrophic/higher salinity waters of the northern Arabian Sea, being found exclusively in the oligotrophic/low salinity southern water mass. It can reasonably be deduced that primary productivity is the main factor determining the distribution of the genetic types across the region, giving G. ruber great potential as a paleoproxy for ocean productivity.
To fulfil this role, a link must be demonstrated between genetic type and subtle variations in shell morphology, as has already been achieved for fellow spinose morphospecies, O. universa [11]. Several morphological variants or 'morphotypes' have already been recorded within G. ruber (white) [76-79] and crucially, G. ruber (white) genetic types I and II can be distinguished morphologically [80], and are consistent with the G. ruber senso stricto (s.s.) and G. ruber senso lato (s.l) morphotypes of Wang [81]. Differences in ecological behaviour have been noted between these two genetic types/morphotypes, with stable isotope and Mg/Ca data together with field observations revealing differing depth habitats and nutrient requirements between the two [78,81-83]. The combined findings suggest an adaptation of G. ruber Type I to oligotrophic, shallow conditions, and an adaptation of Type II to eutrophic, deeper conditions.
In the Arabian Sea we indeed see a clear ecological distinction between the G. ruber Type Ia and Type II lineages; Type 1a occupying only the oligotrophic southern water mass, and Type IIa almost exclusively occupying the more eutrophic north. Type 1a has also been found to be restricted to oligotrophic waters in both the North Atlantic subtropical gyre and the eastern Mediterranean Sea [31]. It likely dominates during the more oligotrophic periods of the seasonal cycle in the Arabian Sea. Of the other Type I genetic types, Ib(1) also fits the 'oligotrophic Type I profile', being present in the low-nutrient south of the Arabian Sea, though it was represented by only a single specimen. Type Ib(2), however, is far from being adapted to oligotrophic conditions, occupying the eutrophic northern water mass of the Arabian Sea together with Type IIa. It seems then that as in G. siphonifera, ecological partitioning of the G. ruber genetic types may not always reflect the Type I and Type II lineage differentiation. Globigerinoides ruber Types Ib(1) and Ib(2) appear ecologically distinct in their distribution patterns, despite being only subtly different at the genetic level.
The three planktonic foraminiferal morphospecies, Globigerina bulloides, Globigerinoides sacculifer, and Orbulina universa, were each represented by only single genetic types in the Arabian Sea, which again displayed non-random biogeographical distributions between the northern and southern water masses. The disparate distributions of Globigerina bulloides and G. sacculifer (Figures 4D, 3 and 4E) give strong indications of specific ecological requirements, which are most likely related to nutrient availability.
Globigerina bulloides is more typical of sub-polar regions [84], but also characterises upwelling zones in lower latitudes [85]. It is comprised of two major lineages (Figure 3C), Type I occurring in warm waters, and Type II occurring in cold waters [9]. In the Arabian Sea only Type Ia was present, occurring predominately towards the north of the region (Figure 3, 4D). It's absence from the most oligotrophic, lower salinity waters (stations 6 - 9) (bulk sample assessment and Figure 3, 4D) perhaps indicates an adaptation to slightly more eutrophic, higher salinity conditions. Interestingly, Globigerina bulloides dominates the planktonic foraminiferal assemblages in the cooler upwelling coastal waters of the Arabian Sea [52]. It remains to be seen whether the warm water genetic type of the central Arabian Sea mixed layer (Type 1a) is ecologically distinct from those found in high numbers in the upwelling coastal regions of the Arabian Sea.
Globigerinoides sacculifer, by contrast, was the dominant morphospecies in the southern Arabian Sea during the SW monsoon. Only a single genetic type of G. sacculifer has been recorded globally. In this study it was confined to the southern oligotrophic water mass (Figure 4E), reflecting a possible adaptation to more oligotrophic waters [86,87]. It has been postulated that other factors such as the chlorophyll maximum or thermocline development may affect its distribution [88], and its status in the Arabian Sea water column has been shown to vary with temperature, salinity, nutrients and thermocline depth [52]. Salinity is an unlikely limiting factor as G. sacculifer is a euryhaline species, capable of tolerating salinities in a range of 24‰ - 47‰ [71].
One further morphospecies, O. universa, was restricted in its distribution. It was represented by only a single Type 1a specimen, found in the oligotrophic southern water mass. Though insufficient data prevents us from drawing conclusions regarding its ecological adaptations, this is consistent with the previous classification of O. universa Type Ia as an oligotrophic-adapted type [11].
Some morphospecies from the Arabian Sea displayed broad distributions during the SW monsoon, indicating that they are not restricted by adaptations to sea surface productivity, the main discriminating ecological factor between the northern & southern water masses. The prominent morphospecies, G. menardii, G. rubescens (pink), N. dutertrei and P. obliquiloculata (Figure 4F, H, I and 4J) were each represented by only single genetic types, exhibiting wide distributions along the whole transect. Bulk samples did indicate that G. menardii numbers tended to increase in the assemblage towards the most southern part of the cruise transect. It should also be noted that different genetic types have potentially been recognised within N. dutertrei [39] and P. obliquiloculata (unpublished observation), though extensive sampling and cloning will be required before their individual biogeographical distributions can be determined.
Other broadly distributed morphospecies included G. glutinata (represented by 2 genetic types), G. ungulata, T. quinqueloba (only one Type 1b specimen genotyped), and S. globigerus (Figures 4C, G, L, and 4M). The tiny spinose morphospecies T. quinqueloba is likely to be underrepresented in the data set; it's small size leading to difficulties in collection. Streptochilus globigerus was found throughout the cruise transect and may be of particular interest. This sporadically occurring, biserial planktonic foraminifer [89,90] displayed such high levels of SSU rDNA sequence identity to the benthic species Bolivina variabilis from the Kenyan coastal region [91] (located south west of our central Arabian Sea sampling stations) that they must represent the same species [72]. In the benthos, B. variabilis/S. globigerus lives as a shallow to intermediate infaunal dweller in the continental shelf sediments. During the SW monsoon (Figure 1B), its populations become expatriated by the winds and currents far offshore, where they continue to live and grow as plankton in the open ocean [72]. Streptochilus globigerus is therefore tychopelagic [92] in nature, exploiting both benthic and planktonic habitats [72].
Ecological processes are increasingly being viewed as a vital mode of diversification in the marine environment, with evidence of ecological partitioning being reported for many marine taxa [4,7-9,20,27-30]. From our study of the tropical Arabian Sea, we have demonstrated that biogeographical distributions of the planktonic foraminiferal morphospecies/genetic types can be influenced by adaptations to differing hydrographic conditions. Salinity is unlikely to be a limiting factor in the biogeographic distributions of planktonic foraminifera, as it has previously been shown that planktonic foraminifera are tolerant of extremes of salinity [93]. We therefore propose that primary productivity is the principal variable determining the disparate distributions observed. Evidence of distinct ecological requirements between closely related genetic types implies that ecological partitioning may indeed play a role in the diversification of some planktonic foraminifera. Vicariant processes, however, have also been shown to play an important role in the diversification of several planktonic foraminiferal morphospecies, particularly in the higher latitudes [6-9,38,39]. In reality, the mechanisms of diversification and speciation within the marine environment are undoubtedly quite complex. It will only be with further research that the relative roles of ecological and vicariant processes can be fully elucidated.
Do algal symbionts play a role in divergent adaptations to sea-surface nutrients?
In the Arabian Sea, primary productivity appeared to represent the primary cause of divergent ecological adaptation amongst foraminiferal genetic types, however, we have yet to explore the biological mechanisms involved. Storz et al. [94] proposed that planktonic foraminiferal species respond primarily to productivity, triggered by the seasonal dynamics of vertical stratification of the upper water column and speculated that the distinct nutrition strategies of strictly asymbiontic, facultatively symbiontic, and symbiontic species may play a key role in explaining their abundances and temporal succession.
In fact differences in symbiont affiliations may indeed help to explain the divergent adaptations to sea-surface nutrients observed in the Arabian Sea morphospecies. Globigerina bulloides, for example, was distributed mainly towards the more eutrophic north of the Arabian Sea and is known to be symbiont barren [71], reliant on high levels of primary productivity and food availability in the water column. Globigerinoides sacculifer conversely was restricted to the oligotrophic waters of the southern water mass, and is known to be obligatory symbiont bearing [71], benefiting from photosynthetic energy contributions. Finally, of the broadly distributed morphospecies within the Arabian Sea, G. menardii, N. dutertrei, P. obliquiloculata, and G. glutinata are all known to harbour facultative symbionts [71], meaning that they can either lack or possess symbionts. This may result in their observed versatility, allowing them to either exploit high nutrient conditions (e.g. those in the northern Arabian Sea), or to survive under highly oligotrophic conditions (e.g. those in the southern Arabian Sea), by means of photosynthesis.
It may be that variations in symbiont association could also be involved in the ecological partitioning of individual genetic types within morphospecies, driving their diversification, though little data is available at present. Differences in symbiotic associations have certainly been cited as a possible explanation for the different depth habits and nutrient requirements of the Type I and Type II lineages in both G. siphonifera [15,75] and G. ruber (corresponding to the G. ruber s.s. or type b "platys" morphotype, and the G. ruber s.l. or type a "normal" morphotype respectively) [79,82]. However, such studies did not account for possible ecological partitioning between genetic sub-types within the major Type I and Type II lineages in either G. siphonifera or G. ruber. The results gained here certainly suggest that not all genetic types fall within the 'Type I' and 'Type II' profiles as far as adaptation to nutrients is concerned. The relationship between ecological partitioning in the planktonic foraminifera genetic types and variations in their symbiotic associations certainly warrants further investigation.
Conclusions
During the SW monsoon, pronounced environmental conditions lead to a strong disparity between the northern and southern water masses of the Arabian Sea. We find a distinct difference in the distribution and ecology of the planktonic foraminifera of the Arabian Sea mixed layer at this time, segregating morphospecies and genetic types between the high salinity, more eutrophic north and the lower salinity, oligotrophic south. In the north, Globigerinoides ruber dominated, followed by Neogloboquadrina dutertrei, Pulleniatina obliquiloculata, Globorotalia menardii, and Globigerinita glutinata. In the south Globigerinoides sacculifer dominated, followed by Globigerinoides ruber and Globorotalia menardii. For those morphospecies represented by complexes of several discrete genetic types within the Arabian Sea mixed layer, individual genetic types were found to have distinct ecologies and novel adaptations to differing physical oceanographic conditions. Globigerinoides ruber showed a clear ecological distinction between its Type Ia/Ib(1) and Type II lineages, supporting past opinions that Types I and II represent independent species [5,9,31]. However, Type Ib(2), did not fit the typical G. ruber Type I 'oligotrophic' profile [79,80,82], indicating a divergent ecological adaptation from close relative, Type Ib(1). Within both Globigerinoides ruber and Globigerinella siphonifera, subtle sub-types were found to display differing geographical distributions, implicating sea-surface productivity as a significant ecological source of divergent selection in closely related planktonic foraminiferal genetic types. Differing symbiotic associations are a possible mechanism by which divergent nutrient-related adaptations may have arisen in the planktonic foraminiferal morphospecies and possibly even their genetic types.
We have found compelling evidence for ecological partitioning within the planktonic foraminifera of the Arabian Sea. Future efforts should now concentrate on gathering similar data from other global locations, to build a more extensive picture of the ecological requirements of the different foraminiferal genetic types. The ability of foraminiferal genetic types to become specialised and adapted to life in regionally distinct ecosystems is a likely driver of their divergence and speciation in the open ocean, running counter to the apparent lack of barriers to gene flow. If the ecologically divergent foraminiferal genetic types could also be identified from the morphology of their shells, it could represent a considerable improvement to quantitative faunal and geochemical palaeoenvironmental reconstructions.
Supplementary Material
Table S1. A list of all planktonic and benthic foraminiferal morphospecies/genetic types included in the phylogenetic analyses, with their GenBank accession numbers.
Figure S1. SSU rDNA sequence alignment for the foraminifera showing the 407 unambiguously aligned nucleotide sites used to reconstruct the main phylogeny in Figure 2.
Figure S2. SSU rDNA sequence alignments for four of the most common Arabian Sea planktonic foraminiferal morphospecies, showing the unambiguously aligned nucleotide sites used to reconstruct the phylogenies in Figure 3: Globigerinella siphonifera and Globigerinella calida (668 bp), Globigerinoides ruber and Globigerinoides conglobatus (589 bp), Globigerina bulloides (669 bp), and Turborotalita quinqueloba (748 bp).
Contributor Information
Heidi A Seears, Email: heidiseears@gmail.com.
Kate F Darling, Email: Kate.Darling@ed.ac.uk.
Christopher M Wade, Email: chris.wade@nottingham.ac.uk.
Acknowledgements
We thank the crew and scientists of RRS Charles Darwin (cruise CD148) for their assistance, Blair Steel for help with sampling during the cruise and Mathew Carroll for assistance with the project. This work was supported by NERC grants NER/J/S33 2000/00860 and NE/D009707/1 (K.F.D) and the BBSRC (PhD studentship BBS/S/K/6 2004/11230). Molecular work was undertaken with support from the Institute of Genetics, University of Nottingham.
Authors' contributions
HAS, KFD and CMW conceived and designed the study. KFD collected the samples, HAS conducted the laboratory work. HAS and CMW analyzed data. HAS, KFD and CMW wrote the manuscript. All authors read and approved the final manuscript.
References
- Finlay BJ. Global Dispersal of Free-Living Microbial Eukaryote Species. Science. 2002;296:1061–1063. doi: 10.1126/science.1070710. [DOI] [PubMed] [Google Scholar]
- Norris RD. Pelagic species diversity, biogeography, and evolution. Paleobiology. 2000;26(4):236–258. doi: 10.1666/0094-8373(2000)26[236:PSDBAE]2.0.CO;2. [DOI] [Google Scholar]
- Palumbi SR. Genetic Divergence, Reproductive Isolation, and Marine Speciation. Annu Rev Ecol Syst. 1994;25:547–572. doi: 10.1146/annurev.es.25.110194.002555. [DOI] [Google Scholar]
- Darling KF, Wade CM, Kroon D, Leigh Brown AJ. Planktic foraminiferal molecular evolution and their polyphyletic origins from benthic taxa. Micropaleontol. 1997;30:251–266. doi: 10.1016/S0377-8398(96)00057-6. [DOI] [Google Scholar]
- Darling KF, Wade CM, Kroon D, Leigh Brown AJ, Bijma J. The diversity and distribution of modern planktic foraminiferal small subunit ribosomal RNA genotypes and their potential as tracers of present and past ocean circulation. Paleoceanography. 1999;14:3–12. doi: 10.1029/1998PA900002. [DOI] [Google Scholar]
- Darling KF, Kucera M, Pudsey CJ, Wade CM. Molecular evidence links cryptic diversification in polar plankton to Quaternary climate dynamics. Proc Natl Acad Sci USA. 2004;101:7657–7662. doi: 10.1073/pnas.0402401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling KF, Kucera M, Kroon D, Wade CM. A resolution for the coiling direction paradox in Neogloboquadrina pachyderma. Paleoceanography. 2006;21:PA2011. doi:10-1029/2005PA001189. [Google Scholar]
- Darling KF, Kucera M, Wade CM. Global molecular phylogeography reveals persistent Arctic circumpolar isolation in a marine planktonic protist. Proc Natl Acad Sci USA. 2007;104:5002–5007. doi: 10.1073/pnas.0700520104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling KF, Wade CM. The genetic diversity of planktic foraminifera and the global distribution of ribosomal RNA genotypes. Mar Micropaleontol. 2008;67:216–238. doi: 10.1016/j.marmicro.2008.01.009. [DOI] [Google Scholar]
- de Vargas C, Zaninetti L, Hilbrecht H, Pawlowski J. Phylogeny and rates of molecular evolution of planktonic foraminifera: SSU rDNA sequences compared to the fossil record. J Mol Evol. 1997;45(3):285–294. doi: 10.1007/PL00006232. [DOI] [PubMed] [Google Scholar]
- de Vargas C, Norris R, Zaninetti L, Gibb SW, Pawlowski J. Molecular evidence of cryptic speciation in planktonic foraminifers and their relation to oceanic provinces. Proc Natl Acad Sci USA. 1999;96:2864–2868. doi: 10.1073/pnas.96.6.2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vargas C, Renaud S, Hilbrecht H, Pawlowski J. Pleistocene adaptive radiation in Globorotalia truncatulinoides: genetic, morphologic, and environmental evidence. Paleobiology. 2001;27(1):104–125. doi: 10.1666/0094-8373(2001)027<0104:PARIGT>2.0.CO;2. [DOI] [Google Scholar]
- de Vargas C, Bonzon M, Rees NW, Pawlowski J, Zaninetti L. A molecular approach to diversity and biogeography in the planktonic foraminifer Globigerinella siphonifera (d'Orbigny) Mar Micropaleontol. 2002;870:1–16. [Google Scholar]
- Goetze E. Global population genetic structure and biogeography of the oceanic copepods Eucalanus hyalinus and E. spinifer. Evolution. 2005;59:2378–2398. [PubMed] [Google Scholar]
- Huber BT, Bijma J, Darling KF. Cryptic speciation in the living planktonic foraminifer Globigerinella siphonifera (d'Orbigny) Paleobiology. 1997;23:33–62. [Google Scholar]
- Saez AG, Probert I, Geisen M, Quinn P, Young JR, Medlin LK. Pseudo-cryptic speciation in coccolithophores. Proc Natl Acad Sci USA. 2003;100:7163–7168. doi: 10.1073/pnas.1132069100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato A, Kooistra WHCF, Levialdi Ghiron JH, Mann DG, Pröschold T, Montresor M. Reproductive Isolation among Sympatric Cryptic Species in Marine Diatoms. Protist. 2007;158:193–207. doi: 10.1016/j.protis.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Kooistra WHCF, Sarno D, Balzano S, Haifeng Gu, Andersen RA, Zingone A. Global diversity and biogeography of Skeletonema species (bacillariophyta) Protist. 2008;159:177–193. doi: 10.1016/j.protis.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Casteleyn G, Leliaert F, Backeljau T, Debeer AE, Kotaki Y, Rhodes L, Lundholm N, Sabbe K, Vyverman W. Limits to gene flow in a cosmopolitan marine planktonic diatom. Proc Natl Acad Sci USA. 2010;107(29):12952–12957. doi: 10.1073/pnas.1001380107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne N, Bonhomme F, David P. Habitat preference and the marine-speciation paradox. Proc R Soc Lond B Biol Sci. 2003;270:1399–1406. doi: 10.1098/rspb.2003.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton PF, Norris RD. Dispersal and biogeography of marine plankton: Long distance dispersal of the foraminifer Truncorotalia truncatulinoides. Geology. 2008;36:899–902. doi: 10.1130/G25232A.1. [DOI] [Google Scholar]
- Darling KF, Wade CM, Steward IA, Kroon D, Dingle R, Leigh Brown AJ. Molecular evidence for genetic mixing of Arctic and Antarctic subpolar populations of planktonic foraminifers. Nature. 2000;405:43–47. doi: 10.1038/35011002. [DOI] [PubMed] [Google Scholar]
- Schluter D. The Ecology of Adaptive Radiation. Oxford: Oxford University Press; 2000. [Google Scholar]
- Schluter D. Ecology and the origin of species. Trends Ecol Evol. 2001;16:372–380. doi: 10.1016/S0169-5347(01)02198-X. [DOI] [PubMed] [Google Scholar]
- Rundle HD, Nosil P. Ecological speciation. Ecol Lett. 2005;8:336–352. doi: 10.1111/j.1461-0248.2004.00715.x. [DOI] [Google Scholar]
- Briggs JC. Proximate sources of marine biodiversity. J Biogeogr. 2006;33:1–10. doi: 10.1111/j.1365-2699.2005.01374.x. [DOI] [Google Scholar]
- Rüber L, Van Tassell JL, Zardoya R. Rapid speciation and ecological divergence in the American seven-spined gobies (Gobiidae, Gobiosomatini) inferred from a molecular phylogeny. Evolution. 2003;57(7):1584–1598. doi: 10.1111/j.0014-3820.2003.tb00366.x. [DOI] [PubMed] [Google Scholar]
- Rocha LA, Robertson DR, Roman J, Bowen BW. Ecological speciation in tropical reef fishes. Proc R Soc Lond B Biol Sci. 2005;272:573–579. doi: 10.1098/2004.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulon E, Not F, Jalabert TC, Massana R, Simon N. Ecological niche partitioning in the picoplanktonic green alga Micromonas pusilla: evidence from environmental surveys using phylogenetic probes. Environ Microbiol. 2008;10(9):2433–2443. doi: 10.1111/j.1462-2920.2008.01673.x. [DOI] [PubMed] [Google Scholar]
- Puebla O. Ecological speciation in marine ν. freshwater fishes. J Fish Biol. 2009;75:960–996. doi: 10.1111/j.1095-8649.2009.02358.x. [DOI] [PubMed] [Google Scholar]
- Aurahs R, Grimm GW, Hemleben V, Hemleben C, Kucera M. Geographical distribution of cryptic genetic types in the planktonic foraminifer Globigerinoides ruber. Mol Ecol. 2009;18:1692–1706. doi: 10.1111/j.1365-294X.2009.04136.x. [DOI] [PubMed] [Google Scholar]
- Darling KF, Kroon D, Wade CM, Leigh Brown AJ. Molecular phylogeny of the planktic foraminifera. J Foraminifer Res. 1996;26(4):324–330. doi: 10.2113/gsjfr.26.4.324. [DOI] [PubMed] [Google Scholar]
- Darling KF, Kroon D, Wade CM, Leigh Brown AJ. In: Microfossils and Oceanic Environments. Moguilevsky A, Whatley RC, editor. Aberystwyth: University of Wales, Aberystwyth Press; 1996. The isolation and amplification of the 18S ribosomal RNA gene from planktonic foraminifers using gametogenic specimens; pp. 249–259. [Google Scholar]
- de Vargas C, Sáez AG, Medlin LK, Thierstein HR. In: Coccolithophores-from molecular processes to global impact. Thierstein HR, Young JR, editor. Springer; 2004. Super-species in the calcareous plankton; pp. 271–298. [Google Scholar]
- Stewart IA, Darling KF, Kroon D, Wade CM, Troelstra SR. Genotypic variability on subarctic Atlantic planktic foraminifera. Mar Micropaleontol. 2001;43:143–153. doi: 10.1016/S0377-8398(01)00024-X. [DOI] [Google Scholar]
- Ujiié Y, Lipps JH. Cryptic diversity in planktic foraminifera in the northwest Pacific Ocean. J Foraminifer Res. 2009;39:145–154. doi: 10.2113/gsjfr.39.3.145. [DOI] [Google Scholar]
- Rutherford S, D'Hondt S, Prell W. Environmental controls on the geographic distribution of zooplankton diversity. Nature. 1999;400:749–753. doi: 10.1038/23449. [DOI] [Google Scholar]
- Kucera M, Darling KF. Cryptic species of planktonic foraminifera: their effect on palaeoceanographic reconstructions. Phil Trans R Soc Lond A. 2002;360:695–718. doi: 10.1098/rsta.2001.0962. [DOI] [PubMed] [Google Scholar]
- Darling KF, Kucera M, Wade CM, von Langen P, Pak D. Seasonal distribution of genetic types of planktonic foraminifer morphospecies in the Santa Barbara Channel and its paleoceanographic implications. Paleoceanography. 2003;18(2):1032–1042. [Google Scholar]
- Brummer GJA, Kroon D. Planktonic foraminifers as tracers of ocean climate history. Amsterdam Free University Press; 1988. pp. 6–346. [Google Scholar]
- Schott FA, McCreary JP Jr. The Monsoon Circulation of the Indian Ocean. Prog Oceanogr. 2001;51:1–123. doi: 10.1016/S0079-6611(01)00083-0. [DOI] [Google Scholar]
- Clemens S, Prell WD, Murray D, Shimmield G, Weedon G. Forcing mechanisms of the Indian Ocean monsoon. Nature. 1991;353:720–725. doi: 10.1038/353720a0. [DOI] [Google Scholar]
- Schott F. Monsoon response of the Somali Current and associated upwelling. Prog Oceanogr. 1983;12:357–381. doi: 10.1016/0079-6611(83)90014-9. [DOI] [Google Scholar]
- Swallow JC. Some Aspects of the Physical Oceanography of the Indian Ocean. Deep-Sea Res A. 1984;31:639–650. doi: 10.1016/0198-0149(84)90032-3. [DOI] [Google Scholar]
- Findlater J. A Major Low-level Air Current Near the Indian Ocean during the Northern Summer. Q J Roy Meteorol Soc. 1996;95:362–380. [Google Scholar]
- Lee C, Jones B, Brink K, Fischer A. The upper-ocean response to monsoonal forcing in the Arabian Sea: seasonal and spatial variability. Deep-Sea Res II. 2000;47:1177–1226. doi: 10.1016/S0967-0645(99)00141-1. [DOI] [Google Scholar]
- Banse K, English DC. Seasonality of coastal zone colour scanner phytoplankton pigment in the offshore oceans. J Geophys Res. 1994;99:7323–7345. doi: 10.1029/93JC02155. [DOI] [Google Scholar]
- Bauer S, Hitchcock G, Olson D. Influence of monsoonally-forced Ekman dynamics upon the surface layer depth and plankton biomass distribution in the Arabian Sea. Deep-Sea Res Pt A. 1991;38:531–553. doi: 10.1016/0198-0149(91)90062-K. [DOI] [Google Scholar]
- Prell WL, Curry WB. Faunal and isotopic indices of monsoonal upwelling: western Arabian Sea. Oceanol Acta. 1981;4:91–98. [Google Scholar]
- Cullen JL, Prell WL. Planktonic Foraminifera of the Northern Indian Ocean: Distribution and preservation in surface sediments. Mar Micropaleontol. 1984;9:1–52. doi: 10.1016/0377-8398(84)90022-7. [DOI] [Google Scholar]
- Kroon D. Distribution of extant planktic foraminiferal assemblages in Red Sea and Northern Indian Ocean surface waters. Rev Espan Micropaleontol. 1991;23(1):37–74. [Google Scholar]
- Schiebel R, Zeltner A, Treppke UF, Waniek JJ, Bollmann J, Rixen T, Hemleben C. Distribution of diatoms, coccolithophores and planktic foraminifers along a trophic gradient during SW monsoon in the Arabian Sea. Mar Micropaleontol. 2004;51(3-4):345–371. doi: 10.1016/j.marmicro.2004.02.001. [DOI] [Google Scholar]
- Prasanna Kumar S, Narvekar J. Seasonal variability of the mixed layer in the central Arabian Sea and its implication on nutrients and primary productivity. Deep-Sea Res II. 2005;52:1848–1861. doi: 10.1016/j.dsr2.2005.06.002. [DOI] [Google Scholar]
- Locarnini RA, Mishonov AV, Antonov JI, Boyer TP, Garcia HE. In: NOAA Atlas NESDIS 61. Washington LS, editor. Washington: GPO; 2006. World Ocean Atlas 2005, Volume 1: Temperature; p. 182. [Google Scholar]
- Antonov JI, Locarnini RA, Boyer TP, Mishonov AV, Garcia HE. In: NOAA Atlas NESDIS 62. Washington LS, editor. U.S: GPO; 2006. World Ocean Atlas 2005, Volume 2: Salinity; p. 182. [Google Scholar]
- Holzmann M, Pawlowski J. Preservation of foraminifera for DNA extraction and PCR amplification. J Foraminifer Res. 1996;26:264–267. doi: 10.2113/gsjfr.26.3.264. [DOI] [Google Scholar]
- Medlin L, Elwood HJ, Stickel S, Sogin ML. Sequence analysis of enzymatically amplified genomic small subunit rRNA genes from the diatom, Skeletonema pustulata. Gene. 1988;71:491–499. doi: 10.1016/0378-1119(88)90066-2. [DOI] [PubMed] [Google Scholar]
- White TJ, Burns T, Lee S, Taylor J. In: PCR protocols: a guide to methods and applications. Innis MA, Gelfland DH, Sninsky JJ, White TJ, editor. San Diego: Harcourt Brace Jovanovich; 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics; pp. 315–322. [Google Scholar]
- Staden R. The Staden Sequence Analysis Package. Mol Biotechnol. 1996;5:233–241. doi: 10.1007/BF02900361. [DOI] [PubMed] [Google Scholar]
- Smith SW, Overbeek R, Woese R, Gilbert W, Gillevet PM. The genetic data environment and expandable GUI for multiple sequence analysis. Comput Appl Biosci. 1994;10:671–675. doi: 10.1093/bioinformatics/10.6.671. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Larget B, Simon DL. Markov chain monte carlo algorithms for the Bayesian analysis of phylogenetic trees. Mol Biol Evol. 1999;16:750–759. doi: 10.1093/oxfordjournals.molbev.a026160. [DOI] [Google Scholar]
- Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Fitch WH. Toward defining the course of evolution: minimum change for a special tree topology. Syst Zool. 1971;20:406–416. doi: 10.2307/2412116. [DOI] [Google Scholar]
- Lanave C, Preparata G, Saccone C, Serio G. A new method for calculating evolutionary substitution rates. J Mol Evol. 1984;20:86–93. doi: 10.1007/BF02101990. [DOI] [PubMed] [Google Scholar]
- Yang Z. Maximum likelihood estimation of phylogeny from DNA sequences when substitution rates vary over sites. Mol Biol Evol. 1993;10:1396–1401. doi: 10.1093/oxfordjournals.molbev.a040082. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52(5):696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sunderland: Sinauer Associates; 1998. [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- Hemleben Ch, Spindler M, Anderson OR, editor. Modern Planktonic Foraminifera. New York: Springer-Verlag; 1989. [Google Scholar]
- Darling KF, Thomas E, Kasemann SA, Seears HA, Smart CW, Wade CM. Surviving mass extinction by bridging the benthic/planktic divide. Proc Natl Acad Sci USA. 2009;106:12629–12633. doi: 10.1073/pnas.0902827106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennett JP, Srinivasan MS, editor. Neogene Planktonic Foraminifera, a Phylogenetic Atlas. Pennsylvania: Hutchinson and Ross; 1983. [Google Scholar]
- Chaisson WP, Pearson PN. In: Proceedings of the Ocean Drilling Program, Scientific Results. 154. Shackleton NJ, Curry WB, Richter C, Bralower TJ, editor. Texas: College Station; 1997. Planktonic foraminifer biostratigraphy at Site 925: Middle Miocene-Pleistocene; pp. 3–32. [Google Scholar]
- Bijma J, Hemleben C, Huber BT, Erlenkeuser H, Kroon D. Experimental determination of the ontogenetic stable isotope variability in two morphotypes of Globigerinella siphonifera (d'Orbigny) Mar Micropaleontol. 1998;35:141–160. doi: 10.1016/S0377-8398(98)00017-6. [DOI] [Google Scholar]
- Saito T, Thompson PR, Breger D. Systematic Index of Recent and Pleistocene Planktonic Foraminifera. Tokyo: University of Tokyo Press; 1981. [Google Scholar]
- Robbins LL, Healy-Williams N. Toward a classification of planktonic-foraminifera based on biochemical, geochemical, and morphological criteria. J Foraminifer Res. 1991;21(2):159–167. doi: 10.2113/gsjfr.21.2.159. [DOI] [Google Scholar]
- Löwemark L, Hong WL, Yui TF, Hung GW. A test of different factors influencing the isotopic signal of planktonic foraminifera in surface sediments from the northern South China Sea. Mar Micropaleontol. 2005;55(1-2):49–62. doi: 10.1016/j.marmicro.2005.02.004. [DOI] [Google Scholar]
- Numberger L, Hemleben C, Hoffmann R, Mackensen A, Schulz H, Wunderlich JM, Kucera M. Habitats, abundance patterns and isotopic signals of morphotypes of the planktonic foraminifer Globigerinoides ruber (d'Orbigny) in the eastern Mediterranean Sea since the Marine Isotopic Stage 12. Mar Micropaleontol. 2009;73:90–104. doi: 10.1016/j.marmicro.2009.07.004. [DOI] [Google Scholar]
- Kuroyanagi A, Tsuchiya M, Kawahata H, Kitazato H. The occurrence of two genotypes of the planktonic foraminifer Globigerinoides ruber (white) and paleoenvironmental implications. Mar Micropaleontol. 2008;68:236–243. doi: 10.1016/j.marmicro.2008.04.004. [DOI] [Google Scholar]
- Wang LJ. Isotopic signals in two morphotypes of Globigerinoides ruber (white) from the South China Sea: implications for monsoon climate change during the last glacial cycle. Palaeogeogr Palaeoclimatol Palaeoecol. 2000;161(3-4):381–394. doi: 10.1016/S0031-0182(00)00094-8. [DOI] [Google Scholar]
- Kuroyanagi A, Kawahata H. Vertical distribution of living planktonic foraminifera in the seas around Japan. Mar Micropaleontol. 2004;53:173–196. doi: 10.1016/j.marmicro.2004.06.001. [DOI] [Google Scholar]
- Steinke S, Chiu HY, Yu PS, Shen CC, Löwemark L, Mii HS, Chen MT. Mg/Ca ratios of two Globigerinoides ruber (white) morphotypes: implications for reconstructing past tropical/subtropical surface water conditions. Geochem Geophys Geosyst. 2005;6:Q11005. doi:10.1029/2005GC000926. [Google Scholar]
- Bé AWH, Tolderlund DS. In: The Micropaleontology of Oceans. Funnell BM, Riedel WR, editor. London: Cambridge University Press; 1971. Distribution and ecology of planktonic foraminifera; pp. 105–150. [Google Scholar]
- Naidu PD, Malmgren BA. A high resolution record of late Quaternary upwelling along the Oman Margin, Arabian Sea based on planktonic foraminifera. Paleoceanography. 1996;11(1):129–140. doi: 10.1029/95PA03198. [DOI] [Google Scholar]
- Halicz E, Reiss Z. Paleoecological relations of Foraminifera in a desert-enclosed sea - The Gulf of Aqaba (Elat), Red Sea. Mar Ecol. 1981;2(1):15–34. doi: 10.1111/j.1439-0485.1981.tb00088.x. [DOI] [Google Scholar]
- Reiss Z, Luz B, Almogi-Labin A, Halicz E, Winter A, Wolf M, Ross DA. Later Quaternary Paleoceanography of the Gulf of Aqaba (Elat), Red Sea. Quat Res. 1980;14:294–308. doi: 10.1016/0033-5894(80)90013-7. [DOI] [Google Scholar]
- Weikert H. In: Key environments: Red Sea. Edwards AJ, Head SM, editor. Oxford: Pergamon Press; 1987. Plankton and the pelagic environment; pp. 90–111. [Google Scholar]
- de Klasz I, Kroon D, Van Hinte JE. Notes on the foraminiferal genera Laterostomella de Klasz and Rerat and Streptochilus Brönnimann and Resig. J Micropaleontol. 1989;8:215–226. doi: 10.1144/jm.8.2.215. [DOI] [Google Scholar]
- Schiebel R, Hemleben C. Modern Planktic Foraminifera. Paläont Z. 2005;79(1):135–148. [Google Scholar]
- Ertan KT, Hemleben V, Hemleben Ch. Molecular evolution of some selected benthic foraminifera as inferred from sequences of the small subunit ribosomal DNA. Mar Micropaleontol. 2004;53:367–388. doi: 10.1016/j.marmicro.2004.08.001. [DOI] [Google Scholar]
- McQuoid MR, Nordberg K. The diatom Paralia sulcata as an environmental indicator species in coastal sediments. Estuar Coast Shelf Sci. 2003;56:339. doi: 10.1016/S0272-7714(02)00187-7. [DOI] [Google Scholar]
- Bijma J, Faber WW Jr, Hemleben C. Temperature and salinity limits for growth and survival of some planktonic foraminifers in laboratory cultures. J Foraminifer Res. 1990;20(2):95–116. doi: 10.2113/gsjfr.20.2.95. [DOI] [Google Scholar]
- Storz D, Schulz H, Waniek JJ, Schulz-Bull DE, Kucera M. Seasonal and interannual variability of the planktic foraminiferal flux in the vicinity of the Azores Current. Deep-Sea Res I. 2009;56(1):107–124. doi: 10.1016/j.dsr.2008.08.009. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. A list of all planktonic and benthic foraminiferal morphospecies/genetic types included in the phylogenetic analyses, with their GenBank accession numbers.
Figure S1. SSU rDNA sequence alignment for the foraminifera showing the 407 unambiguously aligned nucleotide sites used to reconstruct the main phylogeny in Figure 2.
Figure S2. SSU rDNA sequence alignments for four of the most common Arabian Sea planktonic foraminiferal morphospecies, showing the unambiguously aligned nucleotide sites used to reconstruct the phylogenies in Figure 3: Globigerinella siphonifera and Globigerinella calida (668 bp), Globigerinoides ruber and Globigerinoides conglobatus (589 bp), Globigerina bulloides (669 bp), and Turborotalita quinqueloba (748 bp).