Fig. 3. PUF-3 specifically binds high affinity sites in pos-1 and spn-4 3’UTRs.
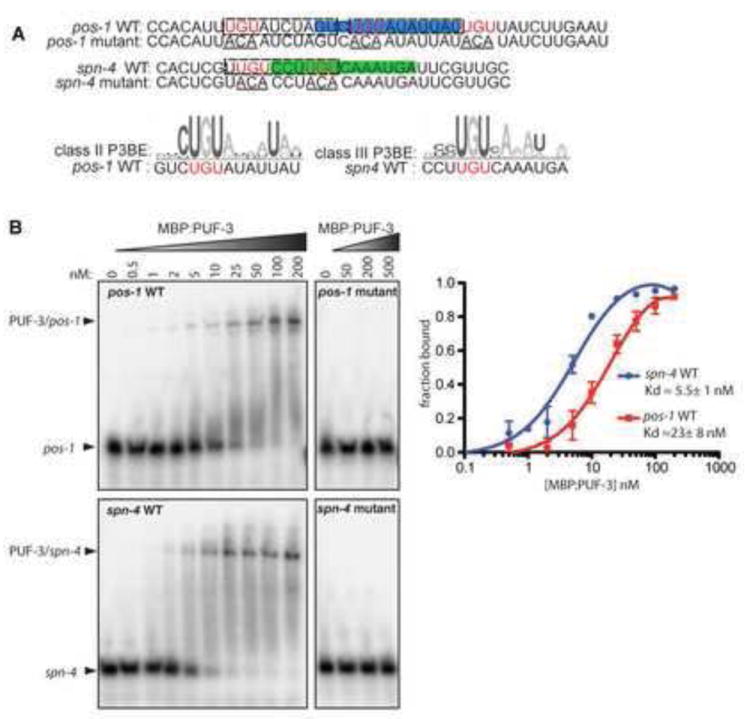
(A) pos-1 and spn-4 3’UTR fragments contain potential PUF-3/11 consensus PBE sequences (P3BEs) consistent with class II (blue) or class III (green) sites, as defined by (Koh et al., 2009). Potential PUF-5 element (P5BE) variants reside in same RNAs (dashed boxes). PBEs in both RNAs are highly conserved (Fig. S1A,B) (B) Gel shift assays of MBP:PUF-3 binding to pos-1 and spn-4 RNAs shown at top of (A). Wild type RNA probes (pos-1 WT and spn-4 WT) contained UGU motifs (red in A) that were mutated to ACA (underlined in A) in pos-1 mutant and spn-4 mutant RNA probes. Arrows indicate unbound RNAs and RNA/PUF-3 complexes. The fraction of bound RNA was plotted against protein concentration to obtain an apparent Kd values.
