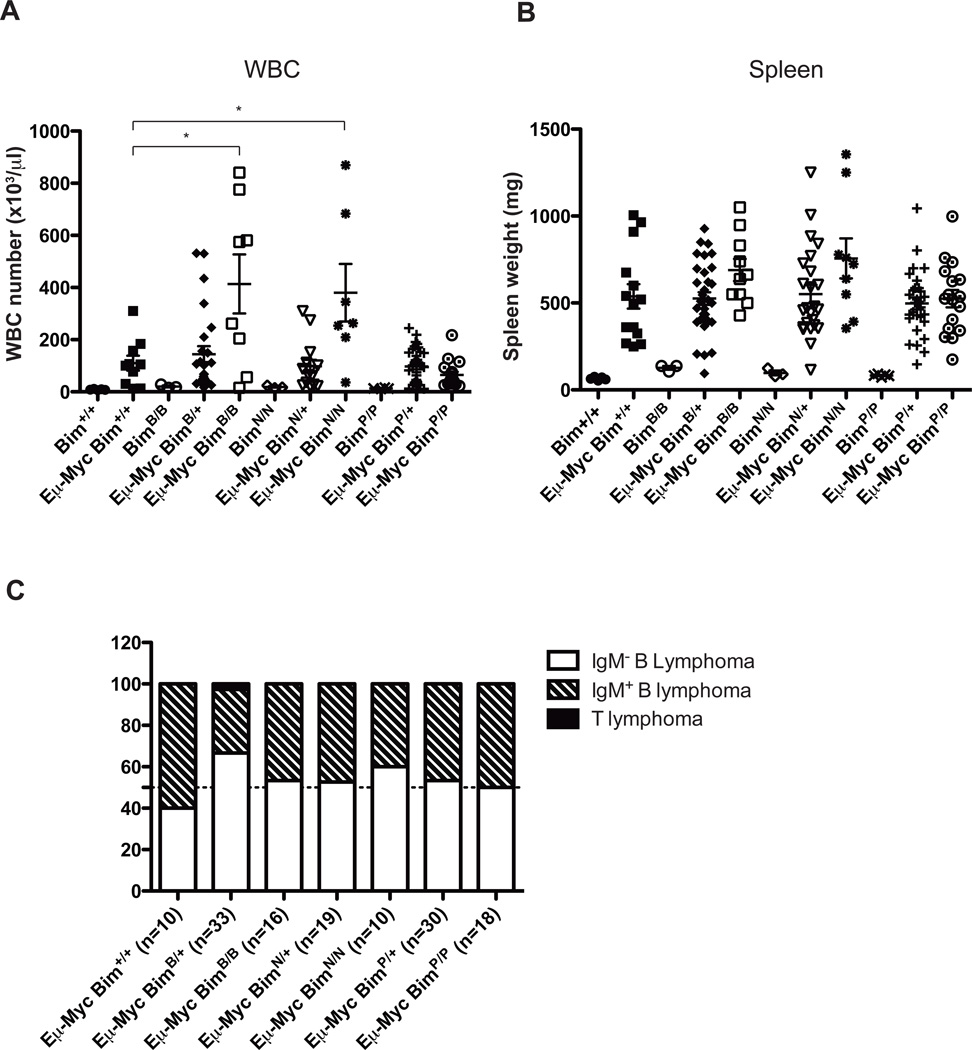
Figure 3. The BimBad and BimNoxa mutations but not the BimPuma mutation increased the leukemic burden in sick Eμ-Myc transgenic mice.
(a) Number of WBC and (b) spleen weight of wt (Bim+/+), control Eμ-Myc (Eμ-Myc/Bim+/+), BimBad/Bad, Eμ-Myc/BimBad/+, Eμ-Myc/BimBad/Bad, BimNoxa/Noxa, Eμ-Myc/BimNoxa/+, Eμ-Myc/BimNoxa/Noxa, BimPuma/Puma, Eμ-Myc/BimPuma/+ and Eμ-Myc/BimPuma/Puma mice at autopsy. Peripheral blood was analyzed using the ADVIA hematology analyzer (Bayer). Data represent means+/−SEM (n=numbers of animals analyzed for each genotype). Statistical analysis was performed using the Student t test. Statistically significant differences in comparison to Eμ-Myc/Bim+/+ mice are indicated (*P<0.05). (c) Diagrammatic representation of the relative frequencies of lymphoma types (sIg−pre-B lymphoma, sIg+ B lymphoma or Thy1+ T lymphoma) observed in mice of the indicated genotypes (n= numbers of mice analyzed for each genotype). Sick mice were sacrificed, lymphomas harvested and single cell suspensions prepared for FACS (FacsCalibur2; Becton Dickinson) analysis using fluorochrome-conjugated antibodies against IgM (5.1 or 333.12), IgD (11-26C) and B220 (RA3-6B2).