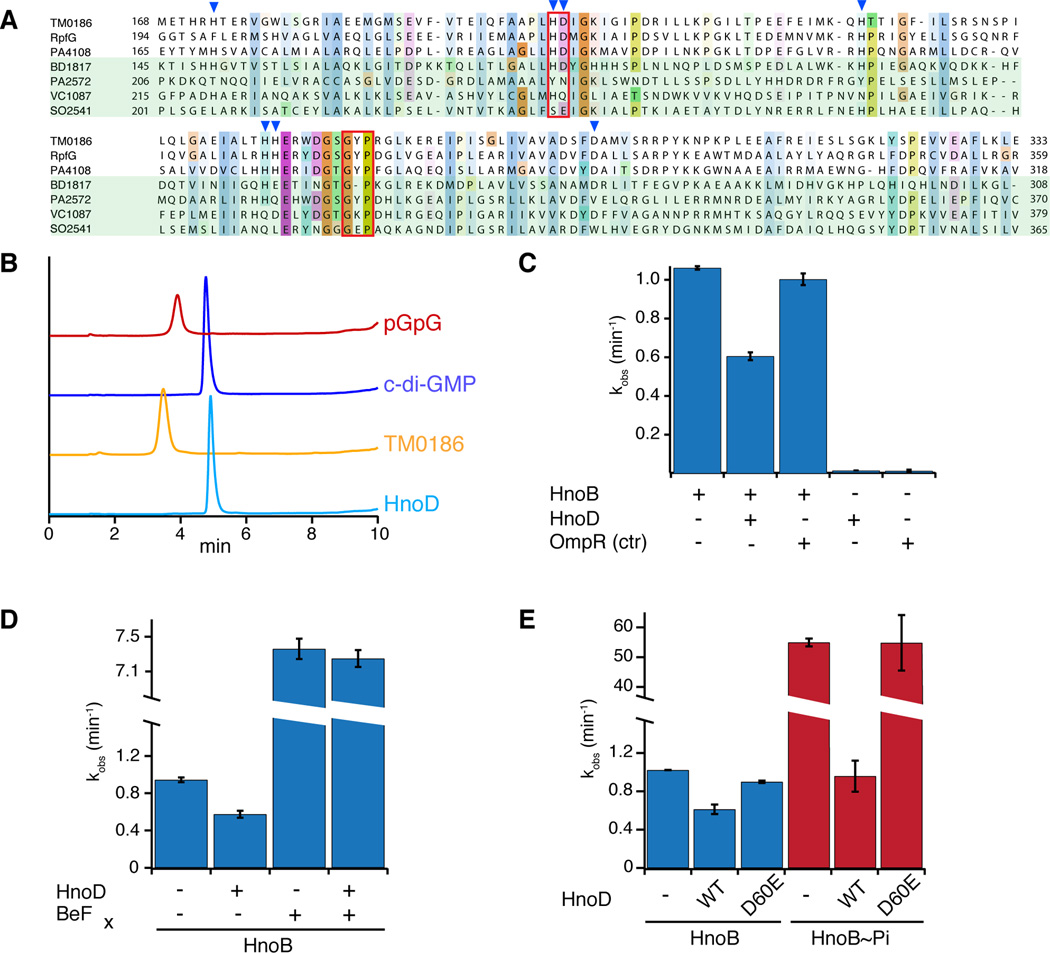
Figure 4. Fine-tuning of HnoB c-di-GMP hydrolysis by the degenerate HD-GYP domain of HnoD.
(A) Sequence alignment of HD-GYP domains from three RRs that contain the conserved sequence motif (T. maritima TM0186, X. campestris RpfG, P. aeruginosa PA4108) and four RR that contain some degeneracy of the conserved sequence (Bdellovibrio BD1817, P. aeruginosa PA2572, V. cholerae HnoD (VC1087), S. oneidensis HnoD (SO2541)). The HD and GYP motifs are boxed in red and residues that are ligands for binuclear iron are indicated by blue triangles. (B) HPLC chromatograms of a PDE assay with a “true” HD-GYP RR (TM0186, yellow) and the degenerate HD-GYP RR (HnoD, light blue) showed that only TM0186 hydrolyzed c-di-GMP to pGpG. Standards of pGpG (red) and cyclic-di-GMP (blue) are shown at the top. (C) HnoD inhibited the PDE activity of the HnoB. Purified HnoB (2.5µM) was pre-incubated with 20x excess HnoD prior to addition of c-di-GMP (0.5mM). (D–E) Phosphorylation of HnoD abolished the inhibition of HnoB PDE activity. (D) Loss of inhibition in the presence of the phosphorylation mimic beryllium fluoride (BeFx). HnoD and HnoB were preincubated with BeFx before the reactions were initiated with c-di-GMP. (E) Inhibition of HnoB by HnoD phosphoacceptor mutants. HnoB was either unphosphorylated (blue) or was pre-phosphorylated with HK (HnoK) and ATP (red). WT HnoD or the phosphorylation mimic D60E mutant was then added prior to initiation of the PDE reaction. All values represent the mean (n = 2) +/− SEM. See also Figure S4.