Figure 4. Proposed ecACP Binding Site on FabB.
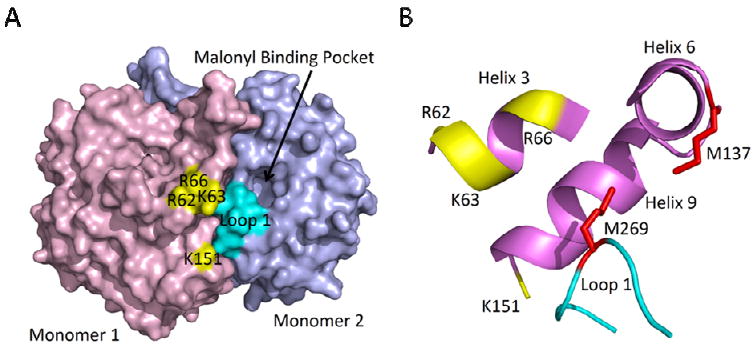
(a) The homodimeric structure of ecFabB (2VB9) in which monomer 1 is colored in purple and monomer 2 is colored in blue. The basic patch that forms the principal ACP binding site in monomer 1 is colored in yellow. R62, K63 and R66 are located on α-helix 3, while K151 is located on α-helix 9 all from monomer 1. Interaction of ACP with this basic patch would result in delivery of the substrate to monomer 2 and interaction of ACP with loop 1 (cyan) in monomer 2. (b) A detailed view of the locations of the conserved methionines that link the active site to helices α5 and α6 are shown in red. They are thought to alter their position upon ecACP binding and aid in the transition from free enzyme to the acyl-enzyme. M269 is located in the middle of loop 1 of monomer 2 and M137 is part of α-helix 6 of monomer 1. The figure was made using PyMOL (52).
