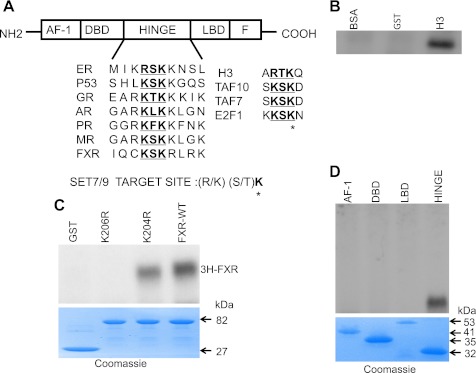
Fig. 6.
Methylation of FXR in vitro by Set7/9 and identification of the lysine methylation site. A: schematic representation of FXR indicating domains conserved in the nuclear hormone receptor superfamily: activation function-1 (AF-1), DNA binding domain (DBD), hinge region (hinge), and ligand-binding domain (LBD). The conserved methylation motif is underlined. The sequence methylated by Set7/9 in various known substrates is indicated to the right. *Target lysine residue. B: strong histone methyltransferase activity of a GST-Set7/9 preparation in vitro is indicated by methylation of recombinant histone H3. GST and BSA were negative control substrates. C: methylation of GST-FXR in an in vitro reaction. GST-Set7/9 (3 μg) was incubated with the GST-fused FXRwt and FXRmutants in the presence of [3H-methyl]-SAM. 3H signal of methylated FXR in the reaction mixture was detected by 10% SDS-PAGE and autoradiography. Recombinant Set7/9 methylated FXR. Mutation of the putative methylation site at K206 (K206R) to an arginine completely abrogated methylation of FXR by Set7/9. FXR was still methylated with mutation of an adjacent, irrelevant lysine (K204) to an arginine. GST lacked Set7/9 activity. bottom: Coomassie blue staining of the gel showing the protein substrates. D: methylation of various GST fused FXR fragments. The hinge domain but not the AF1, DBD, or LBD was methylated by Set7/9. Bottom: Coomassie blue staining of the gel shows the protein substrates.