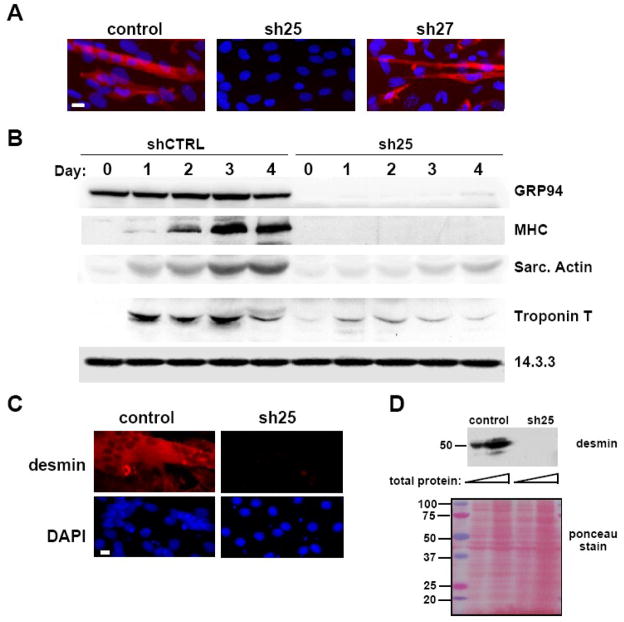
Figure 2. GRP94 silencing results in lack of myogenic differentiation.
A. C2C12 cells transfected either with empty vector (control), or with shRNA25 (sh25) or shRNA27 (sh27) constructs were grown in the presence of serum and induced to differentiate by serum depletion. 48 hrs later, the cells were fixed and stained with the anti-myosin heavy chain antibody MF-20 followed by rhodamine-anti-mouse Ig and also counterstained with DAPI (blue) to visualize nuclei. All cells shown were cultured at the same density. Note the syncitium of nuclei within a fused myotube in the control cells. Magnification bar, 20μm. N=4.
B. C2C12 cells infected with lentiviruses encoding shRNA25 (sh25) or control shRNA (shCTRL) were grown in the presence of serum for 5 days after viral infection. They were then placed in differentiation medium (2% horse serum) (day 0) for the indicated times and equal aliquots of cells were harvested for immunoblots on each of days 0-4. Fifty μg of total protein was loaded on each gel lane. Blots were probed for expression of GRP94. Differentiation was assessed by the induced expression of myosin heavy chain (MHC), sarcomeric actin or troponin T. A representative experiment out of three is shown.
C. C2C12 cells transfected either with empty vector or with shRNA25 were induced to differentiate and 48hs later stained with anti-desmin antibody and rhodamine-anti-rabbit secondary antibody (red, top panels). DAPI staining was used (blue, bottom panels) to highlight nuclei. Magnification bar represents 20μm. N=2.
D. Equal amounts of detergent lysates of the same cultures as in C (15 and 50 μg each sample) were resolved by SDS-PAGE and probed with anti-desmin antibody, to show a second muscle differentiation marker. The membrane stained with ponceau S is shown in the bottom panel, not only to serve as a loading control, but also to show that the patterns of total cell proteins were not affected by the RNAi.