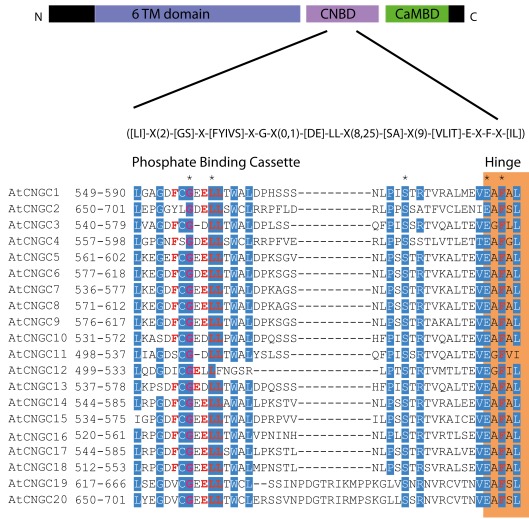
Figure 5.
The A. thaliana CNGC-specific motif spans the putative PBC and the hinge within the CNBD of the 20 CNGCs. The diagram at the top portrays three regions of plant CNGCs: the six transmembrane domains (TM), a CNBD containing a PBC and the hinge which presumably make direct contact with the cyclic nucleotide, and a calmodulin binding domain (CaMBD) toward the C-terminus. The CNGC-specific amino acid motif is shown below the cartoon. In the square brackets “[]” are the amino acids allowed in this position of the motif, “X” represents any amino acid and the round brackets “()” indicate the number of amino acids. The red amino acids in the PBC and the hinge are conserved in both animal and plant CNBDs based on comparison with Jackson et al. (2007). Below the CNGC-specific motif is an alignment of the motif regions of the 20 A. thaliana CNGCs. For each of the A. thaliana CNGCs, the residues at the N- and C-termini are indicated to the left of the motif. The green shaded box demarcates the putative PBC region of the CNBDs (corresponding to the animal CNBD PBC). The orange shaded box marks the presumed hinge region. Residues in white highlighted in blue indicate >90% identity among the 20 A. thaliana CNGCs (in this case “N” is counted as a dissimilarity) indicating a high level of conservation. A “*” above the alignment marks a position with 100% conservation between the A. thaliana sequences shown. Residues in red denote conservation with the animal CNBD alignment generated by Jackson et al. (2007). Note, there is experimental evidence identifying the residues which interact with cyclic nucleotides in animal CNBDs according to Jackson et al., but there is no empirical confirmation of which residues in plant CNBDs bind cyclic nucleotides. The alignment of the A. thaliana CNGC-specific motif was generated by the MEGA5 program and ClustalW (Saitou and Nei, 1987).